2025
|
Noguerol, Julie; Laviolette, Karl; Zahm, Margot; Chaubet, Adeline; Sahal, Ambrine; Détraves, Claire; Torres, Romain; Demont, Clothilde; Adoue, Véronique; Joffre, Carine; Cammas, Florence; van Meerwijk, Joost PM; Joffre, Olivier P. Heterochromatic gene silencing controls CD4+ T cell susceptibility to regulatory T cell-mediated suppression in a murine allograft model Article de journal Dans: Nat Commun, vol. 16, no. 1, 2025, ISSN: 2041-1723. @article{Noguerol2025,
title = {Heterochromatic gene silencing controls CD4+ T cell susceptibility to regulatory T cell-mediated suppression in a murine allograft model},
author = {Julie Noguerol and Karl Laviolette and Margot Zahm and Adeline Chaubet and Ambrine Sahal and Claire Détraves and Romain Torres and Clothilde Demont and Véronique Adoue and Carine Joffre and Florence Cammas and Joost PM van Meerwijk and Olivier P. Joffre},
doi = {10.1038/s41467-025-55848-4},
issn = {2041-1723},
year = {2025},
date = {2025-12-00},
journal = {Nat Commun},
volume = {16},
number = {1},
publisher = {Springer Science and Business Media LLC},
abstract = {AbstractProtective immune responses require close interactions between conventional (Tconv) and regulatory T cells (Treg). The extracellular mediators and signaling events that regulate the crosstalk between these CD4+ T cell subsets have been extensively characterized. However, how Tconv translate Treg-dependent suppressive signals at the chromatin level remains largely unknown. Here we show, using a murine bone marrow allograft model in which graft rejection is coordinated by CD4+ T cells and can be inhibited by Treg, that Treg-mediated T cell suppression involves Heterochromatin Protein 1 α (HP1α)-dependent gene silencing. Unexpectedly, our screen also reveals that T cells deficient for HP1γ or the methyltransferase SUV39H1 are better repressed by Treg than their wild-type counterparts. Mechanistically, our transcriptional and epigenetic profiling identifies HP1γ as a negative regulator of a gene network functionally associated with T-cell exhaustion, including those encoding the inhibitory receptors PD-1 and LAG-3. In conclusion, we identify HP1 variants as rheostats that finely tune the balance between tolerance and immunity. While HP1α converts immunosuppressive signals into heterochromatin-dependent gene silencing mechanisms, HP1γ adjusts Tconv sensitivity to inhibitory environmental signals.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
AbstractProtective immune responses require close interactions between conventional (Tconv) and regulatory T cells (Treg). The extracellular mediators and signaling events that regulate the crosstalk between these CD4+ T cell subsets have been extensively characterized. However, how Tconv translate Treg-dependent suppressive signals at the chromatin level remains largely unknown. Here we show, using a murine bone marrow allograft model in which graft rejection is coordinated by CD4+ T cells and can be inhibited by Treg, that Treg-mediated T cell suppression involves Heterochromatin Protein 1 α (HP1α)-dependent gene silencing. Unexpectedly, our screen also reveals that T cells deficient for HP1γ or the methyltransferase SUV39H1 are better repressed by Treg than their wild-type counterparts. Mechanistically, our transcriptional and epigenetic profiling identifies HP1γ as a negative regulator of a gene network functionally associated with T-cell exhaustion, including those encoding the inhibitory receptors PD-1 and LAG-3. In conclusion, we identify HP1 variants as rheostats that finely tune the balance between tolerance and immunity. While HP1α converts immunosuppressive signals into heterochromatin-dependent gene silencing mechanisms, HP1γ adjusts Tconv sensitivity to inhibitory environmental signals. |
Talpin, Alice; Maia, Ana; Carpier, Jean-Marie; Kulakowski, Guillaume; Aubergeon, Lucie; Kervevan, Jerome; Gaal, Camille; Strozzi, Francesco; Billerey, Coline; Amable, Ludivine; Mersceman, Tifanny; Garnier, Alexandrine; Oliveira, Càtia; Calderon, Carolina; Bachrouche, Diana; Ventujol, Chloé; Bernard, Léa; Manteau, Amandine; Martinez, Jennifer; Bonnet, Michaël; Noguerol, Julie; Laviolette, Karl; Boullerot, Laura; Malfroy, Marine; Chevalier, Gregoire; Adotevi, Olivier; Joffre, Olivier; Idbaih, Ahmed; Vieito, Maria; Ghiringhelli, Francois; Stradella, Agostina; Tabatabai, Ghazaleh; Burger, Michael C; Mildenberger, Iris; Herrlinger, Ulrich; Reardon, David A.; Wick, Wolfgang; Gouttefangeas, Cecile; Bonny, Christophe; Chene, Laurent; Magalhaes, Joao Gamelas Mimicry-based strategy between human and commensal antigens for the development of a new family of immune therapies for cancer Article de journal Dans: J Immunother Cancer, vol. 13, no. 2, 2025, ISSN: 2051-1426. @article{Talpin2025,
title = {Mimicry-based strategy between human and commensal antigens for the development of a new family of immune therapies for cancer},
author = {Alice Talpin and Ana Maia and Jean-Marie Carpier and Guillaume Kulakowski and Lucie Aubergeon and Jerome Kervevan and Camille Gaal and Francesco Strozzi and Coline Billerey and Ludivine Amable and Tifanny Mersceman and Alexandrine Garnier and Càtia Oliveira and Carolina Calderon and Diana Bachrouche and Chloé Ventujol and Léa Bernard and Amandine Manteau and Jennifer Martinez and Michaël Bonnet and Julie Noguerol and Karl Laviolette and Laura Boullerot and Marine Malfroy and Gregoire Chevalier and Olivier Adotevi and Olivier Joffre and Ahmed Idbaih and Maria Vieito and Francois Ghiringhelli and Agostina Stradella and Ghazaleh Tabatabai and Michael C Burger and Iris Mildenberger and Ulrich Herrlinger and David A. Reardon and Wolfgang Wick and Cecile Gouttefangeas and Christophe Bonny and Laurent Chene and Joao Gamelas Magalhaes},
doi = {10.1136/jitc-2024-010192},
issn = {2051-1426},
year = {2025},
date = {2025-02-00},
journal = {J Immunother Cancer},
volume = {13},
number = {2},
publisher = {BMJ},
abstract = {BackgroundMolecular mimicry between commensal bacterial antigens and tumor-associated antigens (TAAs) has shown potential in enhancing antitumor immune responses. This study leveraged this concept using commensal bacterial antigens, termed OncoMimics, to induce TAA-derived peptide (TAAp)-specific cross-reactive cytotoxic T cells and improve the efficacy of peptide-based immunotherapies.MethodsThe discovery of OncoMimics primarily relied on a bioinformatics approach to identify commensal bacteria-derived peptide sequences mimicking TAAps. Several OncoMimics peptide (OMP) candidates were selected in silico based on multiple key parameters to assess their potential to elicit and ameliorate immune responses against TAAs. Selected OMPs were synthesized and tested for their affinity and stability on the major histocompatibility complex (MHC) in vitro and for their capacity to elicit cross-reactive OMP-specific/TAAp-specific CD8+T cell responses in human leukocyte antigen (HLA)-A2-humanized mice, human peripheral blood mononuclear cells (PBMC) and patients with cancer.ResultsSelected OMPs demonstrated superior HLA-A2 binding affinities and stabilities compared with homologous TAAps. Vaccination of HLA-A2-humanized mice with OMPs led to the expansion of OMP-specific CD8+T cells that recognize both OMPs and homologous TAAps, exhibiting cytotoxic capacities towards tumor antigens and resulting in tumor protection in a prophylactic setting. Using PBMCs from HLA-A2+healthy donors, we confirmed the ability of OMPs to elicit potent cross-reactive OMP-specific/TAAp-specific CD8+T-cell responses. Interestingly, we observed a high prevalence of OMP-specific T cells across donors. Cytotoxicity assays revealed that OMP-stimulated human T cells specifically targeted and killed tumor cells loaded with OMPs or TAAps. Preliminary data from an ongoing clinical trial (NCT04116658) support these findings, indicating that OMPs elicit robust OMP-specific/TAAp-specific CD8+T cell responses in patients. Initial immunomonitoring data revealed sustained T-cell responses over time, with T cells maintaining a polyfunctional, cytotoxic and memory phenotype, which is critical for effective antitumor activity and long-term immune surveillance.ConclusionsThese findings suggest that leveraging naturally occurring commensal-derived antigens through OMPs could significantly remodel the tumor immune landscape, offering guidance for a promising strategy for cancer peptide-based immunotherapies.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
BackgroundMolecular mimicry between commensal bacterial antigens and tumor-associated antigens (TAAs) has shown potential in enhancing antitumor immune responses. This study leveraged this concept using commensal bacterial antigens, termed OncoMimics, to induce TAA-derived peptide (TAAp)-specific cross-reactive cytotoxic T cells and improve the efficacy of peptide-based immunotherapies.MethodsThe discovery of OncoMimics primarily relied on a bioinformatics approach to identify commensal bacteria-derived peptide sequences mimicking TAAps. Several OncoMimics peptide (OMP) candidates were selected in silico based on multiple key parameters to assess their potential to elicit and ameliorate immune responses against TAAs. Selected OMPs were synthesized and tested for their affinity and stability on the major histocompatibility complex (MHC) in vitro and for their capacity to elicit cross-reactive OMP-specific/TAAp-specific CD8+T cell responses in human leukocyte antigen (HLA)-A2-humanized mice, human peripheral blood mononuclear cells (PBMC) and patients with cancer.ResultsSelected OMPs demonstrated superior HLA-A2 binding affinities and stabilities compared with homologous TAAps. Vaccination of HLA-A2-humanized mice with OMPs led to the expansion of OMP-specific CD8+T cells that recognize both OMPs and homologous TAAps, exhibiting cytotoxic capacities towards tumor antigens and resulting in tumor protection in a prophylactic setting. Using PBMCs from HLA-A2+healthy donors, we confirmed the ability of OMPs to elicit potent cross-reactive OMP-specific/TAAp-specific CD8+T-cell responses. Interestingly, we observed a high prevalence of OMP-specific T cells across donors. Cytotoxicity assays revealed that OMP-stimulated human T cells specifically targeted and killed tumor cells loaded with OMPs or TAAps. Preliminary data from an ongoing clinical trial (NCT04116658) support these findings, indicating that OMPs elicit robust OMP-specific/TAAp-specific CD8+T cell responses in patients. Initial immunomonitoring data revealed sustained T-cell responses over time, with T cells maintaining a polyfunctional, cytotoxic and memory phenotype, which is critical for effective antitumor activity and long-term immune surveillance.ConclusionsThese findings suggest that leveraging naturally occurring commensal-derived antigens through OMPs could significantly remodel the tumor immune landscape, offering guidance for a promising strategy for cancer peptide-based immunotherapies. |
2024
|
Maire, Kilian; Chamy, Léa; Ghazali, Samira; Carratala-Lasserre, Manon; Zahm, Margot; Bouisset, Clément; Métais, Arnaud; Combes-Soia, Lucie; Fuente-Vizuete, Lidia; Trad, Hussein; Chaubet, Adeline; Savignac, Magali; de Peredo, Anne Gonzalez; Subramaniam, Arun; Joffre, Olivier; Lutz, Pierre G.; Lamsoul, Isabelle Fine-tuning levels of filamins a and b as a specific mechanism sustaining Th2 lymphocyte functions Article de journal Dans: Nature Communications, vol. 15, no. 1, p. 10574, 2024, ISSN: 2041-1723. @article{maire_fine-tuning_2024,
title = {Fine-tuning levels of filamins a and b as a specific mechanism sustaining Th2 lymphocyte functions},
author = {Kilian Maire and Léa Chamy and Samira Ghazali and Manon Carratala-Lasserre and Margot Zahm and Clément Bouisset and Arnaud Métais and Lucie Combes-Soia and Lidia Fuente-Vizuete and Hussein Trad and Adeline Chaubet and Magali Savignac and Anne Gonzalez de Peredo and Arun Subramaniam and Olivier Joffre and Pierre G. Lutz and Isabelle Lamsoul},
doi = {10.1038/s41467-024-53768-3},
issn = {2041-1723},
year = {2024},
date = {2024-12-01},
urldate = {2024-12-01},
journal = {Nature Communications},
volume = {15},
number = {1},
pages = {10574},
abstract = {Augmenting the portfolio of therapeutics for type 2-driven diseases is crucial to address unmet clinical needs and to design personalized treatment schemes. An attractive therapy for such diseases would consist in targeting the recruitment of T helper 2 (Th2) lymphocytes to inflammatory sites. Herein, we show the degradation of filamins (FLN) a and b by the ASB2α E3 ubiquitin ligase as a mechanism sustaining Th2 lymphocyte functions. Low levels of FLNa and FLNb confer an elongated shape to Th2 lymphocytes associated with efficient αVβ3 integrin-dependent cell migration. Genes encoding the αVβ3 integrin and ASB2α belong to the core of Th2-specific genes. Using genetically modified mice, we find that increasing the levels of FLNa and FLNb in Th2 lymphocytes reduces airway inflammation through diminished Th2 lymphocyte recruitment in inflamed lungs. Collectively, our results highlight ASB2α and its substrates FLNa and FLNb to alter Th2 lymphocyte-mediated responses.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
Augmenting the portfolio of therapeutics for type 2-driven diseases is crucial to address unmet clinical needs and to design personalized treatment schemes. An attractive therapy for such diseases would consist in targeting the recruitment of T helper 2 (Th2) lymphocytes to inflammatory sites. Herein, we show the degradation of filamins (FLN) a and b by the ASB2α E3 ubiquitin ligase as a mechanism sustaining Th2 lymphocyte functions. Low levels of FLNa and FLNb confer an elongated shape to Th2 lymphocytes associated with efficient αVβ3 integrin-dependent cell migration. Genes encoding the αVβ3 integrin and ASB2α belong to the core of Th2-specific genes. Using genetically modified mice, we find that increasing the levels of FLNa and FLNb in Th2 lymphocytes reduces airway inflammation through diminished Th2 lymphocyte recruitment in inflamed lungs. Collectively, our results highlight ASB2α and its substrates FLNa and FLNb to alter Th2 lymphocyte-mediated responses. |
Morandi, Elena; Adoue, Véronique; Bernard, Isabelle; Friebel, Ekaterina; Nunez, Nicolas; Aubert, Yann; Masson, Frederick; Dejean, Anne S.; Becher, Burkhard; Astier, Anne; Martinet, Ludovic; Saoudi, Abdelhadi Impact of the Multiple Sclerosis-Associated Genetic Variant
CD226
Gly307Ser on Human CD8 T-Cell Functions Article de journal Dans: Neurol Neuroimmunol Neuroinflamm, vol. 11, no. 6, 2024, ISSN: 2332-7812. @article{Morandi2024,
title = {Impact of the Multiple Sclerosis-Associated Genetic Variant
\textit{CD226}
Gly307Ser on Human CD8 T-Cell Functions},
author = {Elena Morandi and Véronique Adoue and Isabelle Bernard and Ekaterina Friebel and Nicolas Nunez and Yann Aubert and Frederick Masson and Anne S. Dejean and Burkhard Becher and Anne Astier and Ludovic Martinet and Abdelhadi Saoudi},
doi = {10.1212/nxi.0000000000200306},
issn = {2332-7812},
year = {2024},
date = {2024-11-00},
journal = {Neurol Neuroimmunol Neuroinflamm},
volume = {11},
number = {6},
publisher = {Ovid Technologies (Wolters Kluwer Health)},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
|
Debesset, Anais; Pilon, Caroline; Meunier, Sylvain; Cuelenaere-Bonizec, Orianne; Richer, Wilfrid; Thiolat, Allan; Houppe, Claire; Ponzo, Matteo; Magnan, Jeanne; Caron, Jonathan; Caudana, Pamela; Boari, Jimena Tosello; Baulande, Sylvain; To, Nhu Han; Salomon, Benoit Laurent; Piaggio, Eliane; Cascone, Ilaria; Cohen, José Laurent TNFR2 blockade promotes antitumoral immune response in PDAC by targeting activated Treg and reducing T cell exhaustion Article de journal Dans: J Immunother Cancer, vol. 12, no. 11, 2024, ISSN: 2051-1426. @article{Debesset2024,
title = {TNFR2 blockade promotes antitumoral immune response in PDAC by targeting activated Treg and reducing T cell exhaustion},
author = {Anais Debesset and Caroline Pilon and Sylvain Meunier and Orianne Cuelenaere-Bonizec and Wilfrid Richer and Allan Thiolat and Claire Houppe and Matteo Ponzo and Jeanne Magnan and Jonathan Caron and Pamela Caudana and Jimena Tosello Boari and Sylvain Baulande and Nhu Han To and Benoit Laurent Salomon and Eliane Piaggio and Ilaria Cascone and José Laurent Cohen},
doi = {10.1136/jitc-2024-008898},
issn = {2051-1426},
year = {2024},
date = {2024-11-00},
journal = {J Immunother Cancer},
volume = {12},
number = {11},
publisher = {BMJ},
abstract = {BackgroundPancreatic ductal adenocarcinoma (PDAC) is one of the most aggressive cancers, highly resistant to standard chemotherapy and immunotherapy. Regulatory T cells (Tregs) expressing tumor necrosis factor α receptor 2 (TNFR2) contribute to immunosuppression in PDAC. Treg infiltration correlates with poor survival and tumor progression in patients with PDAC. We hypothesized that TNFR2 inhibition using a blocking monoclonal antibody (mAb) could shift the Treg-effector T cell balance in PDAC, thus enhancing antitumoral responses.MethodTo support this hypothesis, we first described TNFR2 expression in a cohort of 24 patients with PDAC from publicly available single-cell analysis data. In orthotopic and immunocompetent mouse models of PDAC, we also described the immune environment of PDAC after immune cell sorting and single-cell analysis. The modifications of the immune environment before and after anti-TNFR2 mAb treatment were evaluated as well as the effect on tumor progression.ResultsPatients with PDAC exhibited elevated TNFR2 expression in Treg, myeloid cells and endothelial cells and lower level in tumor cells. By flow cytometry and single-cell RNA-seq analysis, we identified two Treg populations in orthotopic mouse models: Resting and activated Tregs. The anti-TNFR2 mAb selectively targeted activated tumor-infiltrating Tregs, reducing T cell exhaustion markers in CD8+T cells. However, anti-TNFR2 treatment alone had limited efficacy in activating CD8+T cells and only slightly reduced the tumor growth. The combination of the anti-TNFR2 mAb with agonistic anti-CD40 mAb promoted stronger T cell activation, tumor growth inhibition, and improved survival and immunological memory in PDAC-bearing mice.ConclusionOur data suggest that combining a CD40 agonist with a TNFR2 antagonist represents a promising therapeutic strategy for patients with PDAC.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
BackgroundPancreatic ductal adenocarcinoma (PDAC) is one of the most aggressive cancers, highly resistant to standard chemotherapy and immunotherapy. Regulatory T cells (Tregs) expressing tumor necrosis factor α receptor 2 (TNFR2) contribute to immunosuppression in PDAC. Treg infiltration correlates with poor survival and tumor progression in patients with PDAC. We hypothesized that TNFR2 inhibition using a blocking monoclonal antibody (mAb) could shift the Treg-effector T cell balance in PDAC, thus enhancing antitumoral responses.MethodTo support this hypothesis, we first described TNFR2 expression in a cohort of 24 patients with PDAC from publicly available single-cell analysis data. In orthotopic and immunocompetent mouse models of PDAC, we also described the immune environment of PDAC after immune cell sorting and single-cell analysis. The modifications of the immune environment before and after anti-TNFR2 mAb treatment were evaluated as well as the effect on tumor progression.ResultsPatients with PDAC exhibited elevated TNFR2 expression in Treg, myeloid cells and endothelial cells and lower level in tumor cells. By flow cytometry and single-cell RNA-seq analysis, we identified two Treg populations in orthotopic mouse models: Resting and activated Tregs. The anti-TNFR2 mAb selectively targeted activated tumor-infiltrating Tregs, reducing T cell exhaustion markers in CD8+T cells. However, anti-TNFR2 treatment alone had limited efficacy in activating CD8+T cells and only slightly reduced the tumor growth. The combination of the anti-TNFR2 mAb with agonistic anti-CD40 mAb promoted stronger T cell activation, tumor growth inhibition, and improved survival and immunological memory in PDAC-bearing mice.ConclusionOur data suggest that combining a CD40 agonist with a TNFR2 antagonist represents a promising therapeutic strategy for patients with PDAC. |
Santamaria, Jérémy C.; Vuillier, Sylvia; Galindo-Albarrán, Ariel O.; Castan, Sarah; Detraves, Claire; Joffre, Olivier P.; Romagnoli, Paola; van Meerwijk, Joost P. M. The type 1 diabetes susceptibility locus Idd5 favours robust neonatal development of highly autoreactive regulatory T cells in the NOD mouse Article de journal Dans: Front. Immunol., vol. 15, 2024, ISSN: 1664-3224. @article{Santamaria2024,
title = {The type 1 diabetes susceptibility locus Idd5 favours robust neonatal development of highly autoreactive regulatory T cells in the NOD mouse},
author = {Jérémy C. Santamaria and Sylvia Vuillier and Ariel O. Galindo-Albarrán and Sarah Castan and Claire Detraves and Olivier P. Joffre and Paola Romagnoli and Joost P. M. van Meerwijk},
doi = {10.3389/fimmu.2024.1358459},
issn = {1664-3224},
year = {2024},
date = {2024-02-09},
journal = {Front. Immunol.},
volume = {15},
publisher = {Frontiers Media SA},
abstract = {Regulatory T lymphocytes expressing the transcription factor Foxp3 (Tregs) play an important role in the prevention of autoimmune diseases and other immunopathologies. Aberrations in Treg-mediated immunosuppression are therefore thought to be involved in the development of autoimmune pathologies, but few have been documented. Recent reports indicated a central role for Tregs developing during the neonatal period in the prevention of autoimmune pathology. We therefore investigated the development of Tregs in neonatal NOD mice, an important animal model for autoimmune type 1 diabetes. Surprisingly, we found that, as compared with seven other commonly studied inbred mouse strains, in neonatal NOD mice, exceptionally large proportions of developing Tregs express high levels of GITR and PD-1. The latter phenotype was previously associated with high Treg autoreactivity in C57BL/6 mice, which we here confirm for NOD animals. The proportions of newly developing GITRhighPD-1+ Tregs rapidly drop during the first week of age. A genome-wide genetic screen indicated the involvement of several diabetes susceptibility loci in this trait. Analysis of a congenic mouse strain confirmed that Idd5 contributes to the genetic control of GITRhighPD-1+ Treg development in neonates. Our data thus demonstrate an intriguing and paradoxical correlation between an idiosyncrasy in Treg development in NOD mice and their susceptibility to type 1 diabetes.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
Regulatory T lymphocytes expressing the transcription factor Foxp3 (Tregs) play an important role in the prevention of autoimmune diseases and other immunopathologies. Aberrations in Treg-mediated immunosuppression are therefore thought to be involved in the development of autoimmune pathologies, but few have been documented. Recent reports indicated a central role for Tregs developing during the neonatal period in the prevention of autoimmune pathology. We therefore investigated the development of Tregs in neonatal NOD mice, an important animal model for autoimmune type 1 diabetes. Surprisingly, we found that, as compared with seven other commonly studied inbred mouse strains, in neonatal NOD mice, exceptionally large proportions of developing Tregs express high levels of GITR and PD-1. The latter phenotype was previously associated with high Treg autoreactivity in C57BL/6 mice, which we here confirm for NOD animals. The proportions of newly developing GITRhighPD-1+ Tregs rapidly drop during the first week of age. A genome-wide genetic screen indicated the involvement of several diabetes susceptibility loci in this trait. Analysis of a congenic mouse strain confirmed that Idd5 contributes to the genetic control of GITRhighPD-1+ Treg development in neonates. Our data thus demonstrate an intriguing and paradoxical correlation between an idiosyncrasy in Treg development in NOD mice and their susceptibility to type 1 diabetes. |
Joulia, Emeline; Michieletto, Michaël F.; Agesta, Arantxa; Peillex, Cindy; Girault, Virginie; Dorze, Anne-Louise Le; Peroceschi, Romain; Bucciarelli, Florence; Szelechowski, Marion; Chaubet, Adeline; Hakim, Nawad; Marrocco, Rémi; Lhuillier, Emeline; Lebeurrier, Manuel; Argüello, Rafael J.; Saoudi, Abdelhadi; Costa, Hicham El; Adoue, Veronique; Walzer, Thierry; Sarry, Jean-Emmanuel; Dejean, Anne S. Eomes-dependent mitochondrial regulation promotes survival of pathogenic CD4+ T cells during inflammation Article de journal Dans: vol. 221, no. 2, 2024, ISSN: 1540-9538. @article{Joulia2024,
title = {Eomes-dependent mitochondrial regulation promotes survival of pathogenic CD4+ T cells during inflammation},
author = {Emeline Joulia and Michaël F. Michieletto and Arantxa Agesta and Cindy Peillex and Virginie Girault and Anne-Louise Le Dorze and Romain Peroceschi and Florence Bucciarelli and Marion Szelechowski and Adeline Chaubet and Nawad Hakim and Rémi Marrocco and Emeline Lhuillier and Manuel Lebeurrier and Rafael J. Argüello and Abdelhadi Saoudi and Hicham El Costa and Veronique Adoue and Thierry Walzer and Jean-Emmanuel Sarry and Anne S. Dejean},
doi = {10.1084/jem.20230449},
issn = {1540-9538},
year = {2024},
date = {2024-02-05},
volume = {221},
number = {2},
publisher = {Rockefeller University Press},
abstract = {The mechanisms whereby Eomes controls tissue accumulation of T cells and strengthens inflammation remain ill-defined. Here, we show that Eomes deletion in antigen-specific CD4+ T cells is sufficient to protect against central nervous system (CNS) inflammation. While Eomes is dispensable for the initial priming of CD4+ T cells, it is required for long-term maintenance of CNS-infiltrating CD4+ T cells. We reveal that the impact of Eomes on effector CD4+ T cell longevity is associated with sustained expression of multiple genes involved in mitochondrial organization and functions. Accordingly, epigenetic studies demonstrate that Eomes supports mitochondrial function by direct binding to either metabolism-associated genes or mitochondrial transcriptional modulators. Besides, the significance of these findings was confirmed in CD4+ T cells from healthy donors and multiple sclerosis patients. Together, our data reveal a new mechanism by which Eomes promotes severity and chronicity of inflammation via the enhancement of CD4+ T cell mitochondrial functions and resistance to stress-induced cell death.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
The mechanisms whereby Eomes controls tissue accumulation of T cells and strengthens inflammation remain ill-defined. Here, we show that Eomes deletion in antigen-specific CD4+ T cells is sufficient to protect against central nervous system (CNS) inflammation. While Eomes is dispensable for the initial priming of CD4+ T cells, it is required for long-term maintenance of CNS-infiltrating CD4+ T cells. We reveal that the impact of Eomes on effector CD4+ T cell longevity is associated with sustained expression of multiple genes involved in mitochondrial organization and functions. Accordingly, epigenetic studies demonstrate that Eomes supports mitochondrial function by direct binding to either metabolism-associated genes or mitochondrial transcriptional modulators. Besides, the significance of these findings was confirmed in CD4+ T cells from healthy donors and multiple sclerosis patients. Together, our data reveal a new mechanism by which Eomes promotes severity and chronicity of inflammation via the enhancement of CD4+ T cell mitochondrial functions and resistance to stress-induced cell death. |
2023
|
Rao, Snigdha N; Zahm, Margot; Casemayou, Audrey; Buleon, Marie; Faguer, Stanislas; Feuillet, Guylène; Iacovoni, Jason S; Joffre, Olivier P; Gonzalez-Fuentes, Ignacio; Lhuillier, Emeline; Martins, Frédéric; Riant, Elodie; Zakaroff-Girard, Alexia; Schanstra, Joost P; Blache, Jean Sébastien Saulnier; Belliere, Julie Single-cell RNA sequencing identifies senescence as therapeutic target in rhabdomyolysis-induced acute kidney injury Article de journal Dans: Nephrol Dial Transplant, 2023, ISSN: 1460-2385. @article{pmid37697719,
title = {Single-cell RNA sequencing identifies senescence as therapeutic target in rhabdomyolysis-induced acute kidney injury},
author = {Snigdha N Rao and Margot Zahm and Audrey Casemayou and Marie Buleon and Stanislas Faguer and Guylène Feuillet and Jason S Iacovoni and Olivier P Joffre and Ignacio Gonzalez-Fuentes and Emeline Lhuillier and Frédéric Martins and Elodie Riant and Alexia Zakaroff-Girard and Joost P Schanstra and Jean Sébastien Saulnier Blache and Julie Belliere},
doi = {10.1093/ndt/gfad199},
issn = {1460-2385},
year = {2023},
date = {2023-09-01},
urldate = {2023-09-01},
journal = {Nephrol Dial Transplant},
abstract = {BACKGROUND: The role of macrophages in the development of rhabdomyolysis induced acute kidney injury (RM-AKI) has been established, but an in-depth understanding of the changes in the immune landscape could help to improve targeted strategies. Whereas senescence is usually associated with chronic kidney processes, we also wished to explore whether senescence could also occur in AKI and whether senolytics could act on immune cells.nnMETHODS: Single-cell RNA sequencing was used in the murine glycerol-induced RM-AKI model to dissect the transcriptomic characteristics of CD45+ live cells sorted from kidneys 2 days after injury. Public datasets from murine AKI models were reanalyzed to explore cellular senescence signature in tubular epithelial cells (TECs). A combination of senolytics (dasatinib and quercetin, DQ) was administered to mice exposed or not to RM-AKI.nnRESULTS: Unsupervised clustering of nearly 17,000 single-cell transcriptomes identified 7 known immune cell clusters. Sub-clustering of the mononuclear phagocyte cells (MPC), revealed 9 distinct cell sub-populations differently modified with RM. One macrophage cluster was particularly interesting since it behaved as a critical node in a trajectory connecting one MCHIIhigh cluster only present in control to 2 MCHIIlow clusters only present in RM-AKI. This critical cluster expressed a senescence gene signature, that was very different from that of the TECs. Senolytic DQ treatment blocked the switch from a F4/80highCD11blow to F4/80lowCD11bhigh phenotype, which correlated with prolonged nephroprotection in RM-AKI.nnCONCLUSIONS: scRNASeq unmasked novel transitional macrophage subpopulation associated with RM-AKI characterized by the activation of cellular senescence processes. This work provides a proof-of-concept that senolytics nephroprotective effects may rely, at least in part, on subtle immune modulation.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
BACKGROUND: The role of macrophages in the development of rhabdomyolysis induced acute kidney injury (RM-AKI) has been established, but an in-depth understanding of the changes in the immune landscape could help to improve targeted strategies. Whereas senescence is usually associated with chronic kidney processes, we also wished to explore whether senescence could also occur in AKI and whether senolytics could act on immune cells.nnMETHODS: Single-cell RNA sequencing was used in the murine glycerol-induced RM-AKI model to dissect the transcriptomic characteristics of CD45+ live cells sorted from kidneys 2 days after injury. Public datasets from murine AKI models were reanalyzed to explore cellular senescence signature in tubular epithelial cells (TECs). A combination of senolytics (dasatinib and quercetin, DQ) was administered to mice exposed or not to RM-AKI.nnRESULTS: Unsupervised clustering of nearly 17,000 single-cell transcriptomes identified 7 known immune cell clusters. Sub-clustering of the mononuclear phagocyte cells (MPC), revealed 9 distinct cell sub-populations differently modified with RM. One macrophage cluster was particularly interesting since it behaved as a critical node in a trajectory connecting one MCHIIhigh cluster only present in control to 2 MCHIIlow clusters only present in RM-AKI. This critical cluster expressed a senescence gene signature, that was very different from that of the TECs. Senolytic DQ treatment blocked the switch from a F4/80highCD11blow to F4/80lowCD11bhigh phenotype, which correlated with prolonged nephroprotection in RM-AKI.nnCONCLUSIONS: scRNASeq unmasked novel transitional macrophage subpopulation associated with RM-AKI characterized by the activation of cellular senescence processes. This work provides a proof-of-concept that senolytics nephroprotective effects may rely, at least in part, on subtle immune modulation. |
Pichler, Andrea C.; Carrié, Nadège; Cuisinier, Marine; Ghazali, Samira; Voisin, Allison; Axisa, Pierre-Paul; Tosolini, Marie; Mazzotti, Céline; Golec, Dominic P.; Maheo, Sabrina; do Souto, Laura; Ekren, Rüçhan; Blanquart, Eve; Lemaitre, Lea; Feliu, Virginie; Joubert, Marie-Véronique; Cannons, Jennifer L.; Guillerey, Camille; Avet-Loiseau, Hervé; Watts, Tania H.; Salomon, Benoit L.; Joffre, Olivier; Grinberg-Bleyer, Yenkel; Schwartzberg, Pamela L.; Lucca, Liliana E.; Martinet, Ludovic TCR-independent CD137 (4-1BB) signaling promotes CD8+-exhausted T cell proliferation and terminal differentiation Article de journal Dans: Immunity, vol. 56, no. 7, p. 1631–1648.e10, 2023, ISSN: 1074-7613. @article{Pichler2023b,
title = {TCR-independent CD137 (4-1BB) signaling promotes CD8+-exhausted T cell proliferation and terminal differentiation},
author = {Andrea C. Pichler and Nadège Carrié and Marine Cuisinier and Samira Ghazali and Allison Voisin and Pierre-Paul Axisa and Marie Tosolini and Céline Mazzotti and Dominic P. Golec and Sabrina Maheo and Laura do Souto and Rüçhan Ekren and Eve Blanquart and Lea Lemaitre and Virginie Feliu and Marie-Véronique Joubert and Jennifer L. Cannons and Camille Guillerey and Hervé Avet-Loiseau and Tania H. Watts and Benoit L. Salomon and Olivier Joffre and Yenkel Grinberg-Bleyer and Pamela L. Schwartzberg and Liliana E. Lucca and Ludovic Martinet},
doi = {10.1016/j.immuni.2023.06.007},
issn = {1074-7613},
year = {2023},
date = {2023-07-00},
journal = {Immunity},
volume = {56},
number = {7},
pages = {1631--1648.e10},
publisher = {Elsevier BV},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
|
2022
|
Apert, Cécile; Galindo-Albarrán, Ariel; Castan, Sarah; Detraves, Claire; Michaud, Héloise; McJannett, Nicola; Haegeman, Bart; Fillatreau, Simon; Malissen, Bernard; Hollander, Georg; Zuklys, Saulius; Santamaria, Jérémy; Joffre, Olivier; Romagnoli, Paola; van Meerwijk, Joost IL-2 and IL-15 drive intrathymic development of distinct periphery-seeding CD4+Foxp3+ regulatory T lymphocytes Article de journal Dans: Frontiers in Immunology, 2022. @article{vanJoost.2022,
title = {IL-2 and IL-15 drive intrathymic development of distinct periphery-seeding CD4+Foxp3+ regulatory T lymphocytes},
author = {Cécile Apert and Ariel Galindo-Albarrán and Sarah Castan and Claire Detraves and Héloise Michaud and Nicola McJannett and Bart Haegeman and Simon Fillatreau and Bernard Malissen and Georg Hollander and Saulius Zuklys and Jérémy Santamaria and Olivier Joffre and Paola Romagnoli and Joost van Meerwijk},
url = {https://www.frontiersin.org/articles/10.3389/fimmu.2022.965303/full},
doi = {10.3389/fimmu.2022.965303},
year = {2022},
date = {2022-09-08},
journal = {Frontiers in Immunology},
abstract = {Development of Foxp3-expressing regulatory T-lymphocytes (Treg) in the thymus is controlled by signals delivered in T-cell precursors via the TCR, co-stimulatory receptors, and cytokine receptors. In absence of IL-2, IL-15 or their receptors, fewer Treg apparently develop in the thymus. However, it was recently shown that a substantial part of thymic Treg are cells that had recirculated from the periphery back to the thymus, troubling interpretation of these results. We therefore reassessed the involvement of IL-2 and IL-15 in the development of Treg, taking into account Treg-recirculation. At the age of three weeks, when in wt and IL-15-deficient (but not in IL-2-deficient) mice substantial amounts of recirculating Treg are present in the thymus, we found similarly reduced proportions of newly developed Treg in absence of IL-2 or IL-15, and in absence of both cytokines even less Treg developed. In neonates, when practically no recirculating Treg were found in the thymus, the absence of IL-2 led to substantially more reduced Treg-development than deficiency in IL-15. IL-2 but not IL-15 modulated the CD25, GITR, OX40, and CD73-phenotypes of the thymus-egress-competent and periphery-seeding Treg-population. Interestingly, IL-2 and IL-15 also modulated the TCR-repertoire expressed by developing Treg. Upon transfer into Treg-less Foxp3sf mice, newly developed Treg from IL-2- (and to a much lesser extent IL-15-) deficient mice suppressed immunopathology less efficiently than wt Treg. Taken together, our results firmly establish important non-redundant quantitative and qualitative roles for IL-2 and, to a lesser extent, IL-15 in intrathymic Treg-development.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
Development of Foxp3-expressing regulatory T-lymphocytes (Treg) in the thymus is controlled by signals delivered in T-cell precursors via the TCR, co-stimulatory receptors, and cytokine receptors. In absence of IL-2, IL-15 or their receptors, fewer Treg apparently develop in the thymus. However, it was recently shown that a substantial part of thymic Treg are cells that had recirculated from the periphery back to the thymus, troubling interpretation of these results. We therefore reassessed the involvement of IL-2 and IL-15 in the development of Treg, taking into account Treg-recirculation. At the age of three weeks, when in wt and IL-15-deficient (but not in IL-2-deficient) mice substantial amounts of recirculating Treg are present in the thymus, we found similarly reduced proportions of newly developed Treg in absence of IL-2 or IL-15, and in absence of both cytokines even less Treg developed. In neonates, when practically no recirculating Treg were found in the thymus, the absence of IL-2 led to substantially more reduced Treg-development than deficiency in IL-15. IL-2 but not IL-15 modulated the CD25, GITR, OX40, and CD73-phenotypes of the thymus-egress-competent and periphery-seeding Treg-population. Interestingly, IL-2 and IL-15 also modulated the TCR-repertoire expressed by developing Treg. Upon transfer into Treg-less Foxp3sf mice, newly developed Treg from IL-2- (and to a much lesser extent IL-15-) deficient mice suppressed immunopathology less efficiently than wt Treg. Taken together, our results firmly establish important non-redundant quantitative and qualitative roles for IL-2 and, to a lesser extent, IL-15 in intrathymic Treg-development. |
2021
|
Ariel; Castan Galindo-Albarran, Sarah; Santamaria The Repertoire of Newly Developing Regulatory T Cells in the Type 1 Diabetes-Prone NOD Mouse Is Very Diverse Article de journal Dans: Diabetes, vol. 70, no. 8, p. 1729-1737, 2021, ISSN: 1939-327X. @article{Galindo-Albarrán2021,
title = {The Repertoire of Newly Developing Regulatory T Cells in the Type 1 Diabetes-Prone NOD Mouse Is Very Diverse },
author = {Galindo-Albarran, Ariel; Castan, Sarah; Santamaria, Jérémy; Joffre, Olivier; Haegeman, Bart; Romagnoli, Paola; Van Meerwijk, Joost},
url = {https://diabetes-diabetesjournals-org.proxy.insermbiblio.inist.fr/content/70/8/1729.long},
doi = {10.2337/db20-1072 },
issn = {1939-327X},
year = {2021},
date = {2021-08-02},
journal = {Diabetes},
volume = {70},
number = {8},
pages = {1729-1737},
abstract = {Regulatory T lymphocytes expressing the forkhead/winged helix transcription factor Foxp3 (Treg) play a vital role in the protection of the organism from autoimmune disease and other immunopathologies. The antigen specificity of Treg plays an important role in their in vivo activity. We therefore assessed the diversity of the T-cell receptors (TCRs) for antigen expressed by Treg newly developed in the thymus of autoimmune type 1 diabetes-prone NOD mice and compared it to the control mouse strain C57BL/6. Our results demonstrate that use of the TCRα and TCRβ variable (V) and joining (J) segments, length of the complementarity determining region (CDR) 3, and the diversity of the TCRα and TCRβ chains are comparable between NOD and C57BL/6 mice. Genetic defects affecting the diversity of the TCR expressed by newly developed Treg therefore do not appear to be involved in the etiology of type 1 diabetes in the NOD mouse.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
Regulatory T lymphocytes expressing the forkhead/winged helix transcription factor Foxp3 (Treg) play a vital role in the protection of the organism from autoimmune disease and other immunopathologies. The antigen specificity of Treg plays an important role in their in vivo activity. We therefore assessed the diversity of the T-cell receptors (TCRs) for antigen expressed by Treg newly developed in the thymus of autoimmune type 1 diabetes-prone NOD mice and compared it to the control mouse strain C57BL/6. Our results demonstrate that use of the TCRα and TCRβ variable (V) and joining (J) segments, length of the complementarity determining region (CDR) 3, and the diversity of the TCRα and TCRβ chains are comparable between NOD and C57BL/6 mice. Genetic defects affecting the diversity of the TCR expressed by newly developed Treg therefore do not appear to be involved in the etiology of type 1 diabetes in the NOD mouse. |
Darrigues, Julie; Santamaria, Jeremy C.; Galindo-Albarrán, Ariel; Robey, Ellen A.; Joffre, Olivier P.; van Meerwijk, Joost P. M.; Romagnoli, Paola Robust intrathymic development of regulatory T cells in young NOD mice is rapidly restrained by recirculating cells Article de journal Dans: European Journal of Immunology, 2021, ISSN: 15214141. @article{Darrigues2020b,
title = {Robust intrathymic development of regulatory T cells in young NOD mice is rapidly restrained by recirculating cells},
author = {Darrigues, Julie and Santamaria, Jeremy C. and Galindo-Albarrán, Ariel and Robey, Ellen A. and Joffre, Olivier P. and van Meerwijk, Joost P.M. and Romagnoli, Paola},
url = {https://pubmed.ncbi.nlm.nih.gov/32730634/},
doi = {10.1002/eji.202048743},
issn = {15214141},
year = {2021},
date = {2021-03-01},
journal = {European Journal of Immunology},
publisher = {Wiley-VCH Verlag},
abstract = {Regulatory T lymphocytes (Treg) play a vital role in the protection of the organism against autoimmune pathology. It is therefore paradoxical that comparatively large numbers of Treg were found in the thymus of type I diabetes-prone NOD mice. The Treg population in the thymus is composed of newly developing cells and cells that had recirculated from the periphery back to the thymus. We here demonstrate that exceptionally large numbers of Treg develop in the thymus of young, but not adult, NOD mice. Once emigrated from the thymus, an unusually large proportion of these Treg is activated in the periphery, which causes a particularly abundant accumulation of recirculating Treg in the thymus. These cells then rapidly inhibit de novo development of Treg. The proportions of developing Treg thus reach levels similar to or lower than those found in most other, type 1 diabetes-resistant, inbred mouse strains. Thus, in adult NOD mice the particularly large Treg-niche is actually composed of mostly recirculating cells and only few newly developing Treg.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
Regulatory T lymphocytes (Treg) play a vital role in the protection of the organism against autoimmune pathology. It is therefore paradoxical that comparatively large numbers of Treg were found in the thymus of type I diabetes-prone NOD mice. The Treg population in the thymus is composed of newly developing cells and cells that had recirculated from the periphery back to the thymus. We here demonstrate that exceptionally large numbers of Treg develop in the thymus of young, but not adult, NOD mice. Once emigrated from the thymus, an unusually large proportion of these Treg is activated in the periphery, which causes a particularly abundant accumulation of recirculating Treg in the thymus. These cells then rapidly inhibit de novo development of Treg. The proportions of developing Treg thus reach levels similar to or lower than those found in most other, type 1 diabetes-resistant, inbred mouse strains. Thus, in adult NOD mice the particularly large Treg-niche is actually composed of mostly recirculating cells and only few newly developing Treg. |
2020
|
Maqbool, Muhammad Ahmad; Pioger, Léo; El Aabidine, Amal Zine; Karasu, Nezih; Molitor, Anne Marie; Dao, Lan T. M.; Charbonnier, Guillaume; van Laethem, Francois; Fenouil, Romain; Koch, Frederic; Lacaud, Georges; Gut, Ivo; Gut, Marta; Amigorena, Sebastian; Joffre, Olivier; Sexton, Thomas; Spicuglia, Salvatore; Andrau, Jean Christophe Alternative Enhancer Usage and Targeted Polycomb Marking Hallmark Promoter Choice during T Cell Differentiation Article de journal Dans: Cell Reports, vol. 32, no. 7, 2020, ISSN: 22111247. @article{Maqbool2020b,
title = {Alternative Enhancer Usage and Targeted Polycomb Marking Hallmark Promoter Choice during T Cell Differentiation},
author = {Maqbool, Muhammad Ahmad and Pioger, Léo and El Aabidine, Amal Zine and Karasu, Nezih and Molitor, Anne Marie and Dao, Lan T.M. and Charbonnier, Guillaume and van Laethem, Francois and Fenouil, Romain and Koch, Frederic and Lacaud, Georges and Gut, Ivo and Gut, Marta and Amigorena, Sebastian and Joffre, Olivier and Sexton, Thomas and Spicuglia, Salvatore and Andrau, Jean Christophe},
url = {https://pubmed.ncbi.nlm.nih.gov/32814051/},
doi = {10.1016/j.celrep.2020.108048},
issn = {22111247},
year = {2020},
date = {2020-08-01},
journal = {Cell Reports},
volume = {32},
number = {7},
publisher = {Elsevier B.V.},
abstract = {Development and activation of T lymphocytes are coordinated by lineage-specific transcriptional programs. Here, Maqbool et al. performed a wide epigenomic and transcriptional analysis of mouse T cell differentiation. These data provide new insights into the role of multiple enhancers and PRC2 in controlling alternative promoter choice during differentiation.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
Development and activation of T lymphocytes are coordinated by lineage-specific transcriptional programs. Here, Maqbool et al. performed a wide epigenomic and transcriptional analysis of mouse T cell differentiation. These data provide new insights into the role of multiple enhancers and PRC2 in controlling alternative promoter choice during differentiation. |
Adoue, Véronique; Joffre, Olivier Endogenous retroviruses: Friend or foe of the immune system? Article de journal Dans: Médecine/Sciences, vol. 36, no. 3, p. 253-260, 2020, ISSN: 19585381. @article{Adoue2020,
title = {Endogenous retroviruses: Friend or foe of the immune system?},
author = {Adoue, Véronique and Joffre, Olivier},
url = {https://pubmed.ncbi.nlm.nih.gov/32228844/},
doi = {10.1051/medsci/2020022},
issn = {19585381},
year = {2020},
date = {2020-03-01},
booktitle = {Medecine/Sciences},
journal = {Médecine/Sciences},
volume = {36},
number = {3},
pages = {253-260},
publisher = {Editions EDK},
abstract = {Upon priming by dendritic cells, naïve CD4 T lymphocytes are exposed to distinct molecular environments depending on the nature of the pathological stimulus. In response, they mobilize different gene networks that establish lineage-specific developmental programs, and coordinate the acquisition of specific phenotype and functions. Accordingly, CD4 T cells are capable of differentiation into a large variety of functionally-distinct T helper (Th) cell subsets. In this review, we describe the molecular events that control CD4 T cell differentiation at the level of the chromatin. We insist on recent works that have highlighted the key role of H3K9me3-dependent epigenetic mechanisms in the regulation of T cell identity. Interestingly, these pathways shape and control the developmental programs at least in part through the regulation of endogenous retroviruses-derived sequences that have been exapted into cis-regulatory modules of Th genes.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
Upon priming by dendritic cells, naïve CD4 T lymphocytes are exposed to distinct molecular environments depending on the nature of the pathological stimulus. In response, they mobilize different gene networks that establish lineage-specific developmental programs, and coordinate the acquisition of specific phenotype and functions. Accordingly, CD4 T cells are capable of differentiation into a large variety of functionally-distinct T helper (Th) cell subsets. In this review, we describe the molecular events that control CD4 T cell differentiation at the level of the chromatin. We insist on recent works that have highlighted the key role of H3K9me3-dependent epigenetic mechanisms in the regulation of T cell identity. Interestingly, these pathways shape and control the developmental programs at least in part through the regulation of endogenous retroviruses-derived sequences that have been exapted into cis-regulatory modules of Th genes. |
2019
|
Gehrmann, Ulf; Burbage, Marianne; Zueva, Elina; Goudot, Christel; Esnault, Cyril; Ye, Mengliang; Carpier, Jean Marie; Burgdorf, Nina; Hoyler, Thomas; Suarez, Guadalupe; Joannas, Leonel; Heurtebise-Chrétien, Sandrine; Durand, Sylvère; Panes, Rébecca; Bellemare-Pelletier, Angélique; Sáez, Pablo J.; Aprahamian, Fanny; Lefevre, Deborah; Adoue, Veronique; El Aabidine, Amal Zine; Ahmad, Maqbool Muhammad; Hivroz, Claire; Joffre, Olivier; Cammas, Florence; Kroemer, Guido; Gagnon, Etienne; Andrau, Jean Christophe; Amigorena, Sebastian Critical role for TRIM28 and HP1beta/gamma in the epigenetic control of T cell metabolic reprograming and effector differentiation Article de journal Dans: Proceedings of the National Academy of Sciences of the United States of America, vol. 116, no. 51, p. 25839–25849, 2019, ISSN: 10916490. @article{Gehrmann2019,
title = {Critical role for TRIM28 and HP1beta/gamma in the epigenetic control of T cell metabolic reprograming and effector differentiation},
author = {Gehrmann, Ulf and Burbage, Marianne and Zueva, Elina and Goudot, Christel and Esnault, Cyril and Ye, Mengliang and Carpier, Jean Marie and Burgdorf, Nina and Hoyler, Thomas and Suarez, Guadalupe and Joannas, Leonel and Heurtebise-Chrétien, Sandrine and Durand, Sylvère and Panes, Rébecca and Bellemare-Pelletier, Angélique and Sáez, Pablo J. and Aprahamian, Fanny and Lefevre, Deborah and Adoue, Veronique and El Aabidine, Amal Zine and Ahmad, Maqbool Muhammad and Hivroz, Claire and Joffre, Olivier and Cammas, Florence and Kroemer, Guido and Gagnon, Etienne and Andrau, Jean Christophe and Amigorena, Sebastian},
url = {https://pubmed.ncbi.nlm.nih.gov/31776254/},
doi = {10.1073/pnas.1901639116},
issn = {10916490},
year = {2019},
date = {2019-12-01},
journal = {Proceedings of the National Academy of Sciences of the United States of America},
volume = {116},
number = {51},
pages = {25839--25849},
publisher = {National Academy of Sciences},
abstract = {Naive CD4+ T lymphocytes differentiate into different effector types, including helper and regulatory cells (Th and Treg, respectively). Heritable gene expression programs that define these effector types are established during differentiation, but little is known about the epigenetic mechanisms that install and maintain these programs. Here, we use mice defective for different components of heterochromatin-dependent gene silencing to investigate the epigenetic control of CD4+ T cell plasticity. We show that, upon T cell receptor (TCR) engagement, naive and regulatory T cells defective for TRIM28 (an epigenetic adaptor for histone binding modules) or for heterochromatin protein 1 $beta$ and $gamma$ isoforms (HP1$beta$/$gamma$, 2 histone-binding factors involved in gene silencing) fail to effectively signal through the PI3K–AKT–mTOR axis and switch to glycolysis. While differentiation of naive TRIM28−/− T cells into cytokine-producing effector T cells is impaired, resulting in reduced induction of autoimmune colitis, TRIM28−/− regulatory T cells also fail to expand in vivo and to suppress autoimmunity effectively. Using a combination of transcriptome and chromatin immunoprecipitation-sequencing (ChIP-seq) analyses for H3K9me3, H3K9Ac, and RNA polymerase II, we show that reduced effector differentiation correlates with impaired transcriptional silencing at distal regulatory regions of a defined set of Treg-associated genes, including, for example, NRP1 or Snai3. We conclude that TRIM28 and HP1$beta$/$gamma$ control metabolic reprograming through epigenetic silencing of a defined set of Treg-characteristic genes, thus allowing effective T cell expansion and differentiation into helper and regulatory phenotypes.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
Naive CD4+ T lymphocytes differentiate into different effector types, including helper and regulatory cells (Th and Treg, respectively). Heritable gene expression programs that define these effector types are established during differentiation, but little is known about the epigenetic mechanisms that install and maintain these programs. Here, we use mice defective for different components of heterochromatin-dependent gene silencing to investigate the epigenetic control of CD4+ T cell plasticity. We show that, upon T cell receptor (TCR) engagement, naive and regulatory T cells defective for TRIM28 (an epigenetic adaptor for histone binding modules) or for heterochromatin protein 1 $beta$ and $gamma$ isoforms (HP1$beta$/$gamma$, 2 histone-binding factors involved in gene silencing) fail to effectively signal through the PI3K–AKT–mTOR axis and switch to glycolysis. While differentiation of naive TRIM28−/− T cells into cytokine-producing effector T cells is impaired, resulting in reduced induction of autoimmune colitis, TRIM28−/− regulatory T cells also fail to expand in vivo and to suppress autoimmunity effectively. Using a combination of transcriptome and chromatin immunoprecipitation-sequencing (ChIP-seq) analyses for H3K9me3, H3K9Ac, and RNA polymerase II, we show that reduced effector differentiation correlates with impaired transcriptional silencing at distal regulatory regions of a defined set of Treg-associated genes, including, for example, NRP1 or Snai3. We conclude that TRIM28 and HP1$beta$/$gamma$ control metabolic reprograming through epigenetic silencing of a defined set of Treg-characteristic genes, thus allowing effective T cell expansion and differentiation into helper and regulatory phenotypes. |
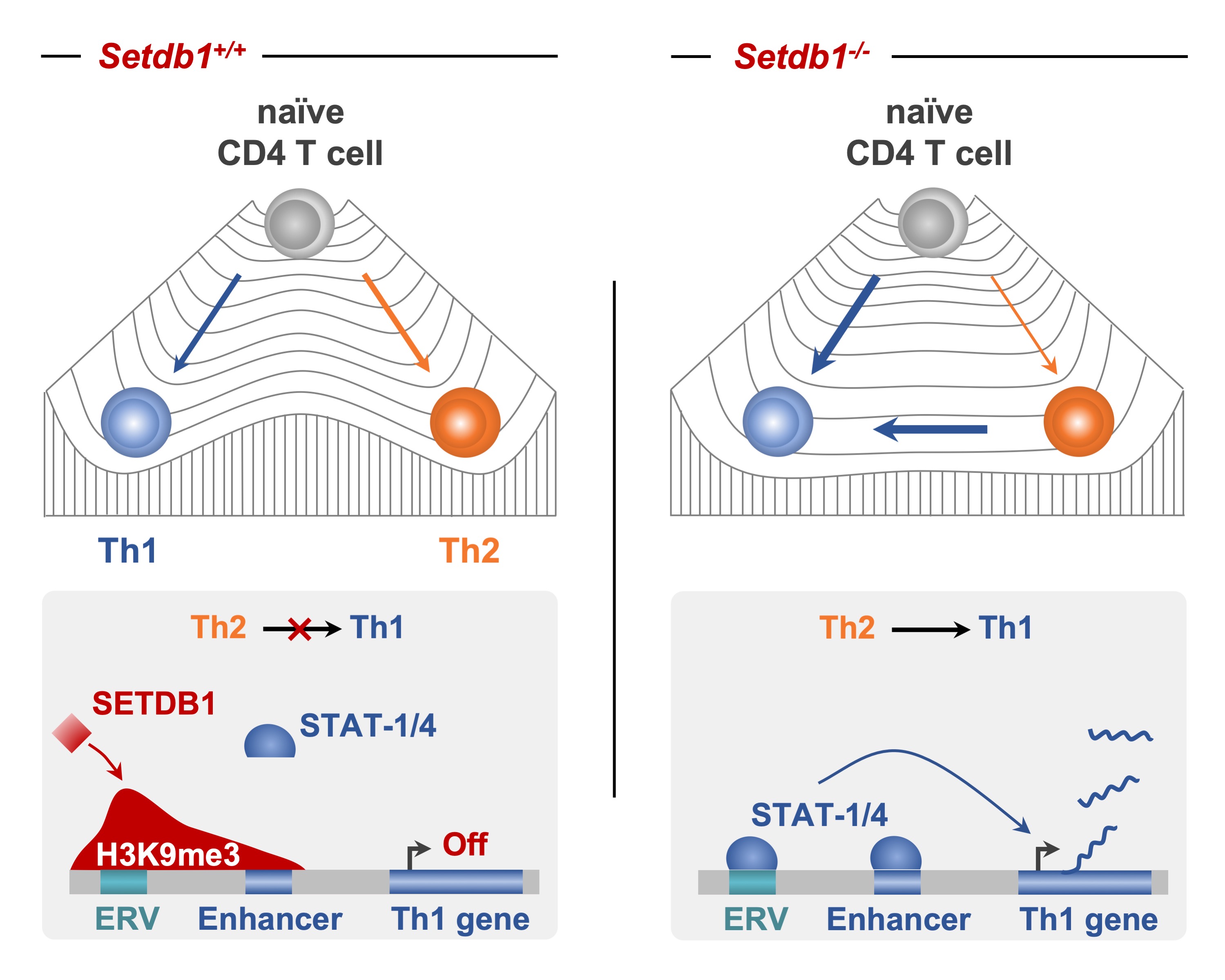
Adoue, V; Binet, B; Malbec, A; Fourquet, J; Romagnoli, P; van Meerwijk, J P M; Amigorena, S; Joffre, O P The Histone Methyltransferase SETDB1 Controls T Helper Cell Lineage Integrity by Repressing Endogenous Retroviruses. Article de journal Dans: Immunity, vol. 50, p. 629–644, 2019. @article{Adoue2019,
title = {The Histone Methyltransferase SETDB1 Controls T Helper Cell Lineage Integrity by Repressing Endogenous Retroviruses.},
author = {Adoue, V and Binet, B and Malbec, A and Fourquet, J and Romagnoli, P and van Meerwijk, J P M and Amigorena, S and Joffre, O P},
url = {https://linkinghub.elsevier.com/retrieve/pii/S1074761319300032},
doi = {10.1016/j.immuni.2019.01.003},
year = {2019},
date = {2019-01-01},
journal = {Immunity},
volume = {50},
pages = {629--644},
abstract = {Upon activation, naive CD4+ T cells differentiate into distinct T cell subsets via processes reliant on epigenetically regulated, lineage-specific developmental programs. Here, we examined the function of the histone methyltransferase SETDB1 in T helper (Th) cell differentiation. Setdb1-/- naive CD4+ T cells exhibited exacerbated Th1 priming, and when exposed to a Th1-instructive signal, Setdb1-/- Th2 cells crossed lineage boundaries and acquired a Th1 phenotype. SETDB1 did not directly control Th1 gene promoter activity but relied instead on deposition of the repressive H3K9me3 mark at a restricted and cell-type-specific set of endogenous retroviruses (ERVs) located in the vicinity of genes involved in immune processes. Refined bioinformatic analyses suggest that these retrotransposons regulate Th1 gene cis-regulatory elements or act as Th1 gene enhancers. Thus, H3K9me3 deposition by SETDB1 ensures Th cell lineage integrity by repressing a repertoire of ERVs that have been exapted into cis-regulatory modules to shape and control the Th1 gene network.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
Upon activation, naive CD4+ T cells differentiate into distinct T cell subsets via processes reliant on epigenetically regulated, lineage-specific developmental programs. Here, we examined the function of the histone methyltransferase SETDB1 in T helper (Th) cell differentiation. Setdb1-/- naive CD4+ T cells exhibited exacerbated Th1 priming, and when exposed to a Th1-instructive signal, Setdb1-/- Th2 cells crossed lineage boundaries and acquired a Th1 phenotype. SETDB1 did not directly control Th1 gene promoter activity but relied instead on deposition of the repressive H3K9me3 mark at a restricted and cell-type-specific set of endogenous retroviruses (ERVs) located in the vicinity of genes involved in immune processes. Refined bioinformatic analyses suggest that these retrotransposons regulate Th1 gene cis-regulatory elements or act as Th1 gene enhancers. Thus, H3K9me3 deposition by SETDB1 ensures Th cell lineage integrity by repressing a repertoire of ERVs that have been exapted into cis-regulatory modules to shape and control the Th1 gene network. |
2018
|
Araujo Furlan, C. L.; Tosello Boari, J.; Rodriguez, C.; Canale, F. P.; Fiocca Vernengo, F.; Boccardo, S.; Beccaria, C. G.; Adoue, V.; Joffre, O.; Gruppi, A.; Montes, C. L.; Acosta Rodriguez, E. V. Limited Foxp3(+) Regulatory T Cells Response During Acute Trypanosoma cruzi Infection Is Required to Allow the Emergence of Robust Parasite-Specific CD8(+) T Cell Immunity Article de journal Dans: Front Immunol, vol. 9, p. 2555, 2018, ISSN: 1664-3224. @article{RN185,
title = {Limited Foxp3(+) Regulatory T Cells Response During Acute Trypanosoma cruzi Infection Is Required to Allow the Emergence of Robust Parasite-Specific CD8(+) T Cell Immunity},
author = {Araujo Furlan, C. L. and Tosello Boari, J. and Rodriguez, C. and Canale, F. P. and Fiocca Vernengo, F. and Boccardo, S. and Beccaria, C. G. and Adoue, V. and Joffre, O. and Gruppi, A. and Montes, C. L. and Acosta Rodriguez, E. V.},
doi = {10.3389/fimmu.2018.02555},
issn = {1664-3224},
year = {2018},
date = {2018-01-01},
journal = {Front Immunol},
volume = {9},
pages = {2555},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
|
Santamaria, J.; Darrigues, J.; van Meerwijk, J. P. M.; Romagnoli, P. Antigen-presenting cells and T-lymphocytes homing to the thymus shape T cell development Article de journal Dans: Immunol Lett, vol. 204, p. 9-15, 2018, ISSN: 0165-2478. @article{RN184,
title = {Antigen-presenting cells and T-lymphocytes homing to the thymus shape T cell development},
author = {Santamaria, J. and Darrigues, J. and van Meerwijk, J. P. M. and Romagnoli, P.},
doi = {10.1016/j.imlet.2018.10.003},
issn = {0165-2478},
year = {2018},
date = {2018-01-01},
journal = {Immunol Lett},
volume = {204},
pages = {9-15},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
|
2017
|
Darrigues, J.; van Meerwijk, J. P. M.; Romagnoli, P. Age-Dependent Changes in Regulatory T Lymphocyte Development and Function: A Mini-Review Article de journal Dans: Gerontology, 2017, ISSN: 0304-324x. @article{RN183,
title = {Age-Dependent Changes in Regulatory T Lymphocyte Development and Function: A Mini-Review},
author = {Darrigues, J. and van Meerwijk, J. P. M. and Romagnoli, P.},
doi = {10.1159/000478044},
issn = {0304-324x},
year = {2017},
date = {2017-01-01},
journal = {Gerontology},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
|
Vuddamalay, Y.; van Meerwijk, J. P. M. CD28neg and CD28low CD8+ regulatory T cells: Of Mice and Men Article de journal Dans: Frontiers in immunology, vol. doi: 10.3389/fimmu.2017.00031, 2017. @article{RN178,
title = {CD28neg and CD28low CD8+ regulatory T cells: Of Mice and Men},
author = {Vuddamalay, Y. and van Meerwijk, J.P.M.},
year = {2017},
date = {2017-01-01},
journal = {Frontiers in immunology},
volume = {doi: 10.3389/fimmu.2017.00031},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
|
Apert, C.; Romagnoli, P.; van Meerwijk, J. P. M. IL-2 and IL-15 dependent thymic development of Foxp3-expressing regulatory T lymphocytes Article de journal Dans: Protein Cell, 2017, ISSN: 1674-800x. @article{RN182b,
title = {IL-2 and IL-15 dependent thymic development of Foxp3-expressing regulatory T lymphocytes},
author = {Apert, C. and Romagnoli, P. and van Meerwijk, J. P. M.},
doi = {10.1007/s13238-017-0425-3},
issn = {1674-800x},
year = {2017},
date = {2017-01-01},
journal = {Protein Cell},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
|
2016
|
Decque, A. *; Joffre, O. *; Magalhaes, J. G. *; Cossec, J. C. *; Blecher-Gonen, R.; Lapaquette, P.; Silvin, A.; Manel, N.; Joubert, P. E.; Seeler, J. S.; Albert, M. L.; Amit, I.; Amigorena, S.; Dejean, A. Sumoylation coordinates the repression of inflammatory and anti-viral gene-expression programs during innate sensing Article de journal Dans: Nat Immunol, vol. 17, no. 2, p. 140-9, 2016, ISSN: 1529-2908. @article{RN181,
title = {Sumoylation coordinates the repression of inflammatory and anti-viral gene-expression programs during innate sensing},
author = {Decque, A.* and Joffre, O.* and Magalhaes, J. G.* and Cossec, J. C.* and Blecher-Gonen, R. and Lapaquette, P. and Silvin, A. and Manel, N. and Joubert, P. E. and Seeler, J. S. and Albert, M. L. and Amit, I. and Amigorena, S. and Dejean, A.},
doi = {10.1038/ni.3342},
issn = {1529-2908},
year = {2016},
date = {2016-01-01},
journal = {Nat Immunol},
volume = {17},
number = {2},
pages = {140-9},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
|
Hamdi, Y; Soucy, P; Adoue, V; Michailidou, K; Canisius, S; Lemaçon, A; Droit, A; Andrulis, I L; Anton-Culver, H; Arndt, V; Baynes, C; Blomqvist, C; Bogdanova, N V; Bojesen, S E; Bolla, M K; Bonanni, B; Borresen-Dale, AL; Brand, J S; Brauch, H; Brenner, H; Broeks, A; Burwinkel, B; Chang-Claude, J; Couch, F J; Cox, A; Cross, S S; Czene, K; Darabi, H; Dennis, J; Devilee, Pr; Dörk, T; Dos-Santos-Silva, I; Eriksson, M; Fasching, P A; Figueroa, J; Flyger, H; Garcia-Closas, M; Giles, G G; Goldberg, M S; Gonzalez-Neira, A; Grenaker-Alnaes, G; Guénel, P; Haeberle, L; Haiman, C A; Hamann, U; Hallberg, E; Hooning, M J; Hopper, J L; Jakubowska, A; Jones, M; Kabisch, M; Kataja, V; Lambrechts, D; Land Lindblom Le Marchand, A; Jand Mannermaa Lubinski, A; Maranian, M; Margolin, S; Marme, F; Milne, R L; Neuhausen, S L; Nevanlinna, H; Neven, P; Olswold, C; Peto, J; Plaseska-Karanfilska, D; Pylkäs, K; Radice, P; Rudolph, A; Sawyer, E J; Schmidt, M K; Shu, X O; Southey, M C; Swerdlow, A; Tollenaar, R A E M; Tomlinson, I; Torres, D; Truong, T; Vachon, C; Van Den Ouweland, A M W; Wang, Q; Winqvist, R; Zheng, W; Benitez, J; Chenevix-Trench, G; Dunning, A M.; Pharoah, P D P; Kristensen, V; Hall, P; Easton, D F; Pastinen, T; Nord, S; Simard, J Association of breast cancer risk with genetic variants showing differential allelic expression: Identification of a novel breast cancer susceptibility locus at 4q21 Article de journal Dans: Oncotarget, 2016, ISSN: 19492553. @article{Hamdi2016,
title = {Association of breast cancer risk with genetic variants showing differential allelic expression: Identification of a novel breast cancer susceptibility locus at 4q21},
author = {Hamdi, Y and Soucy, P and Adoue, V and Michailidou, K and Canisius, S and Lemaçon, A and Droit, A and Andrulis, I L and Anton-Culver, H and Arndt, V and Baynes, C and Blomqvist, C and Bogdanova, N V and Bojesen, S E and Bolla, M K and Bonanni, B and Borresen-Dale, AL and Brand, J S and Brauch, H and Brenner, H and Broeks, A and Burwinkel, B and Chang-Claude, J and Couch, F J and Cox, A and Cross, S S and Czene, K and Darabi, H and Dennis, J and Devilee, Pr and Dörk, T and Dos-Santos-Silva, I and Eriksson, M and Fasching, P A and Figueroa, J and Flyger, H and Garcia-Closas, M and Giles, G G and Goldberg, M S and Gonzalez-Neira, A and Grenaker-Alnaes, G and Guénel, P and Haeberle, L and Haiman, C A and Hamann, U and Hallberg, E and Hooning, M J and Hopper, J L and Jakubowska, A and Jones, M and Kabisch, M and Kataja, V and Lambrechts, D and Le Marchand, Land Lindblom, A and Lubinski, Jand Mannermaa, A and Maranian, M and Margolin, S and Marme, F and Milne, R L and Neuhausen, S L and Nevanlinna, H and Neven, P and Olswold, C and Peto, J and Plaseska-Karanfilska, D and Pylkäs, K and Radice, P and Rudolph, A and Sawyer, E J and Schmidt, M K and Shu, X O and Southey, M C and Swerdlow, A and Tollenaar, R A E M and Tomlinson, I and Torres, D and Truong, T and Vachon, C and Van Den Ouweland, A M W and Wang, Q and Winqvist, R and Zheng, W and Benitez, J and Chenevix-Trench, G and Dunning, A M. and Pharoah, P D P and Kristensen, V and Hall, P and Easton, D F and Pastinen, T and Nord, S and Simard, J},
doi = {10.18632/oncotarget.12818},
issn = {19492553},
year = {2016},
date = {2016-01-01},
journal = {Oncotarget},
abstract = {There are significant inter-individual differences in the levels of gene expression. Through modulation of gene expression, cis-acting variants represent an important source of phenotypic variation. Consequently, cis-regulatory SNPs associated with differential allelic expression are functional candidates for further investigation as disease-causing variants. To investigate whether common variants associated with differential allelic expression were involved in breast cancer susceptibility, a list of genes was established on the basis of their involvement in cancer related pathways and/or mechanisms. Thereafter, using data from a genome-wide map of allelic expression associated SNPs, 313 genetic variants were selected and their association with breast cancer risk was then evaluated in 46,451 breast cancer cases and 42,599 controls of European ancestry ascertained from 41 studies participating in the Breast Cancer Association Consortium. The associations were evaluated with overall breast cancer risk and with estrogen receptor negative and positive disease. One novel breast cancer susceptibility locus on 4q21 (rs11099601) was identified (OR = 1.05},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
There are significant inter-individual differences in the levels of gene expression. Through modulation of gene expression, cis-acting variants represent an important source of phenotypic variation. Consequently, cis-regulatory SNPs associated with differential allelic expression are functional candidates for further investigation as disease-causing variants. To investigate whether common variants associated with differential allelic expression were involved in breast cancer susceptibility, a list of genes was established on the basis of their involvement in cancer related pathways and/or mechanisms. Thereafter, using data from a genome-wide map of allelic expression associated SNPs, 313 genetic variants were selected and their association with breast cancer risk was then evaluated in 46,451 breast cancer cases and 42,599 controls of European ancestry ascertained from 41 studies participating in the Breast Cancer Association Consortium. The associations were evaluated with overall breast cancer risk and with estrogen receptor negative and positive disease. One novel breast cancer susceptibility locus on 4q21 (rs11099601) was identified (OR = 1.05 |
Vuddamalay, Y.; Attia, M.; Vicente, R.; Pomie, C.; Enault, G.; Leobon, B.; Joffre, O.; Romagnoli, P.; van Meerwijk, J. P. Mouse and human CD8(+) CD28(low) regulatory T lymphocytes differentiate in the thymus Article de journal Dans: Immunology, vol. 148, no. 2, p. 187-96, 2016, ISSN: 1365-2567 (Electronic)
0019-2805 (Linking). @article{RN177,
title = {Mouse and human CD8(+) CD28(low) regulatory T lymphocytes differentiate in the thymus},
author = {Vuddamalay, Y. and Attia, M. and Vicente, R. and Pomie, C. and Enault, G. and Leobon, B. and Joffre, O. and Romagnoli, P. and van Meerwijk, J. P.},
url = {http://www.ncbi.nlm.nih.gov/pubmed/26924728},
doi = {10.1111/imm.12600},
issn = {1365-2567 (Electronic)
0019-2805 (Linking)},
year = {2016},
date = {2016-01-01},
journal = {Immunology},
volume = {148},
number = {2},
pages = {187-96},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
|
2015
|
Thiault, N.; Darrigues, J.; Adoue, V.; Gros, M.; Binet, B.; Perals, C.; Leobon, B.; Fazilleau, N.; Joffre, O. P.; Robey, E. A.; van Meerwijk, J. P. M.; Romagnoli, P. Peripheral regulatory T lymphocytes recirculating to the thymus suppress the development of their precursors Article de journal Dans: Nature Immunol, vol. 16, p. 628–634, 2015. @article{RN176,
title = {Peripheral regulatory T lymphocytes recirculating to the thymus suppress the development of their precursors},
author = {Thiault, N. and Darrigues, J. and Adoue, V. and Gros, M. and Binet, B. and Perals, C. and Leobon, B. and Fazilleau, N. and Joffre, O.P. and Robey, E.A. and van Meerwijk, J.P.M. and Romagnoli, P.},
doi = {10.1038/ni.3150},
year = {2015},
date = {2015-01-01},
journal = {Nature Immunol},
volume = {16},
pages = {628–634},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
|
2014
|
Adoue, V; Schiavi, A; Light, N; Almlof, J C; Lundmark, P; Ge, B; Kwan, T; Caron, M; Ronnblom, L; Wang, C; Chen, S H; Goodall, A H; Cambien, F; Deloukas, P; Ouwehand, W H; Syvanen, A C; Pastinen, T Allelic expression mapping across cellular lineages to establish impact of non-coding SNPs Article de journal Dans: Molecular Systems Biology, vol. 10, no. 10, p. 754, 2014. @article{Adoue2014,
title = {Allelic expression mapping across cellular lineages to establish impact of non-coding SNPs},
author = {Adoue, V and Schiavi, A and Light, N and Almlof, J C and Lundmark, P and Ge, B and Kwan, T and Caron, M and Ronnblom, L and Wang, C and Chen, S H and Goodall, A H and Cambien, F and Deloukas, P and Ouwehand, W H and Syvanen, A C and Pastinen, T},
url = {http://msb.embopress.org/cgi/doi/10.15252/msb.20145114},
doi = {10.15252/msb.20145114},
year = {2014},
date = {2014-01-01},
journal = {Molecular Systems Biology},
volume = {10},
number = {10},
pages = {754},
publisher = {EMBO Press},
abstract = {Most complex disease‐associated genetic variants are located in non‐coding regions and are therefore thought to be regulatory in nature. Association mapping of differential allelic expression (AE) is a powerful method to identify SNPs with direct cis‐ regulatory impact ( cis‐ rSNPs). We used AE mapping to identify cis‐ rSNPs regulating gene expression in 55 and 63 HapMap lymphoblastoid cell lines from a Caucasian and an African population, respectively, 70 fibroblast cell lines, and 188 purified monocyte samples and found 40–60% of these cis ‐rSNPs to be shared across cell types. We uncover a new class of cis ‐rSNPs, which disrupt footprint‐derived de novo motifs that are predominantly bound by repressive factors and are implicated in disease susceptibility through overlaps with GWAS SNPs. Finally, we provide the proof‐of‐principle for a new approach for genome‐wide functional validation of transcription factor–SNP interactions. By perturbing NF$kappa$B action in lymphoblasts, we identified 489 cis ‐regulated transcripts with altered AE after NF$kappa$B perturbation. Altogether, we perform a comprehensive analysis of cis ‐variation in four cell populations and provide new tools for the identification of functional variants associated to complex diseases.![][1]textless/imgtextgreaterA comprehensive analysis cis ‐variation in four cell‐populations uncovers a new‐class of cis ‐regulatory SNPs associated with repressor activity. Global analysis of the role of key regulators is achieved by combining allelic expression read‐outs with targeted perturbation of transcription factors.Mol Syst Biol. (2014) 10: 754 [1]: pending:yes},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
Most complex disease‐associated genetic variants are located in non‐coding regions and are therefore thought to be regulatory in nature. Association mapping of differential allelic expression (AE) is a powerful method to identify SNPs with direct cis‐ regulatory impact ( cis‐ rSNPs). We used AE mapping to identify cis‐ rSNPs regulating gene expression in 55 and 63 HapMap lymphoblastoid cell lines from a Caucasian and an African population, respectively, 70 fibroblast cell lines, and 188 purified monocyte samples and found 40–60% of these cis ‐rSNPs to be shared across cell types. We uncover a new class of cis ‐rSNPs, which disrupt footprint‐derived de novo motifs that are predominantly bound by repressive factors and are implicated in disease susceptibility through overlaps with GWAS SNPs. Finally, we provide the proof‐of‐principle for a new approach for genome‐wide functional validation of transcription factor–SNP interactions. By perturbing NF$kappa$B action in lymphoblasts, we identified 489 cis ‐regulated transcripts with altered AE after NF$kappa$B perturbation. Altogether, we perform a comprehensive analysis of cis ‐variation in four cell populations and provide new tools for the identification of functional variants associated to complex diseases.![][1]textless/imgtextgreaterA comprehensive analysis cis ‐variation in four cell‐populations uncovers a new‐class of cis ‐regulatory SNPs associated with repressor activity. Global analysis of the role of key regulators is achieved by combining allelic expression read‐outs with targeted perturbation of transcription factors.Mol Syst Biol. (2014) 10: 754 [1]: pending:yes |
Light, N; Adoue, V; Ge, B; Chen, S-H; Kwan, T; Pastinen, T Interrogation of allelic chromatin states in human cells by high-density ChIP-genotyping. Article de journal Dans: Epigenetics, vol. 9, no. 9, p. 1238–1251, 2014. @article{Light2014,
title = {Interrogation of allelic chromatin states in human cells by high-density ChIP-genotyping.},
author = {Light, N and Adoue, V and Ge, B and Chen, S-H and Kwan, T and Pastinen, T},
url = {http://eutils.ncbi.nlm.nih.gov/entrez/eutils/elink.fcgi?dbfrom=pubmed&id=25055051&retmode=ref&cmd=prlinks},
doi = {10.4161/epi.29920},
year = {2014},
date = {2014-01-01},
journal = {Epigenetics},
volume = {9},
number = {9},
pages = {1238--1251},
abstract = {Allele-specific (AS) assessment of chromatin has the potential to elucidate specific cis-regulatory mechanisms, which are predicted to underlie the majority of the known genetic associations to complex disease. However, development of chromatin landscapes at allelic resolution has been challenging since sites of variable signal strength require substantial read depths not commonly applied in sequencing based approaches. In this study, we addressed this by performing parallel analyses of input DNA and chromatin immunoprecipitates (ChIP) on high-density Illumina genotyping arrays. Allele-specificity for the histone modifications H3K4me1, H3K4me3, H3K27ac, H3K27me3, and H3K36me3 was assessed using ChIP samples generated from 14 lymphoblast and 6 fibroblast cell lines. AS-ChIP SNPs were combined into domains and validated using high-confidence ChIP-seq sites. We observed characteristic patterns of allelic-imbalance for each histone-modification around allele-specifically expressed transcripts. Notably, we found H3K4me1 to be significantly anti-correlated with allelic expression (AE) at transcription start sites, indicating H3K4me1 allelic imbalance as a marker of AE. We also found that allelic chromatin domains exhibit population and cell-type specificity as well as heritability within trios. Finally, we observed that a subset of allelic chromatin domains is regulated by DNase I-sensitive quantitative trait loci and that these domains are significantly enriched for genome-wide association studies hits, with autoimmune disease associated SNPs specifically enriched in lymphoblasts. This study provides the first genome-wide maps of allelic-imbalance for five histone marks. Our results provide new insights into the role of chromatin in cis-regulation and highlight the need for high-depth sequencing in ChIP-seq studies along with the need to improve allele-specificity of ChIP-enrichment.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
Allele-specific (AS) assessment of chromatin has the potential to elucidate specific cis-regulatory mechanisms, which are predicted to underlie the majority of the known genetic associations to complex disease. However, development of chromatin landscapes at allelic resolution has been challenging since sites of variable signal strength require substantial read depths not commonly applied in sequencing based approaches. In this study, we addressed this by performing parallel analyses of input DNA and chromatin immunoprecipitates (ChIP) on high-density Illumina genotyping arrays. Allele-specificity for the histone modifications H3K4me1, H3K4me3, H3K27ac, H3K27me3, and H3K36me3 was assessed using ChIP samples generated from 14 lymphoblast and 6 fibroblast cell lines. AS-ChIP SNPs were combined into domains and validated using high-confidence ChIP-seq sites. We observed characteristic patterns of allelic-imbalance for each histone-modification around allele-specifically expressed transcripts. Notably, we found H3K4me1 to be significantly anti-correlated with allelic expression (AE) at transcription start sites, indicating H3K4me1 allelic imbalance as a marker of AE. We also found that allelic chromatin domains exhibit population and cell-type specificity as well as heritability within trios. Finally, we observed that a subset of allelic chromatin domains is regulated by DNase I-sensitive quantitative trait loci and that these domains are significantly enriched for genome-wide association studies hits, with autoimmune disease associated SNPs specifically enriched in lymphoblasts. This study provides the first genome-wide maps of allelic-imbalance for five histone marks. Our results provide new insights into the role of chromatin in cis-regulation and highlight the need for high-depth sequencing in ChIP-seq studies along with the need to improve allele-specificity of ChIP-enrichment. |
2013
|
Tellier, J.; Andrianjaka, A.; Vicente, R.; Thiault, N.; Enault, G.; Garchon, H. J.; van Meerwijk, J. P. M.; Romagnoli, P. Increased thymic development of regulatory T cells in NOD mice is functionally dissociated from type I diabetes susceptibility Article de journal Dans: Eur J Immunol, vol. 43, no. 5, p. 1356-62, 2013, ISSN: 1521-4141 (Electronic)
0014-2980 (Linking). @article{RN174,
title = {Increased thymic development of regulatory T cells in NOD mice is functionally dissociated from type I diabetes susceptibility},
author = {Tellier, J. and Andrianjaka, A. and Vicente, R. and Thiault, N. and Enault, G. and Garchon, H. J. and van Meerwijk, J. P. M. and Romagnoli, P.},
url = {http://www.ncbi.nlm.nih.gov/pubmed/23400928},
doi = {10.1002/eji.201243142},
issn = {1521-4141 (Electronic)
0014-2980 (Linking)},
year = {2013},
date = {2013-01-01},
journal = {Eur J Immunol},
volume = {43},
number = {5},
pages = {1356-62},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
|
Pasquet, L.; Douet, J. Y.; Sparwasser, T.; Romagnoli, P.; van Meerwijk, J. P. M. Long-term prevention of chronic allograft rejection by regulatory T-cell immunotherapy involves host Foxp3-expressing T cells Article de journal Dans: Blood, vol. 121, no. 21, p. 4303-10, 2013, ISSN: 1528-0020 (Electronic)
0006-4971 (Linking). @article{RN173,
title = {Long-term prevention of chronic allograft rejection by regulatory T-cell immunotherapy involves host Foxp3-expressing T cells},
author = {Pasquet, L. and Douet, J. Y. and Sparwasser, T. and Romagnoli, P. and van Meerwijk, J. P. M.},
url = {http://www.ncbi.nlm.nih.gov/pubmed/23580663},
doi = {10.1182/blood-2012-08-452037},
issn = {1528-0020 (Electronic)
0006-4971 (Linking)},
year = {2013},
date = {2013-01-01},
journal = {Blood},
volume = {121},
number = {21},
pages = {4303-10},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
|
2012
|
Romagnoli, P.; Dooley, J.; Enault, G.; Vicente, R.; Malissen, B.; Liston, A.; van Meerwijk, J. P. M. The thymic niche does not limit development of the naturally diverse population of mouse regulatory T lymphocytes Article de journal Dans: Journal of Immunology, vol. 189, no. 8, p. 3831-7, 2012, ISSN: 1550-6606 (Electronic)
0022-1767 (Linking). @article{RN166,
title = {The thymic niche does not limit development of the naturally diverse population of mouse regulatory T lymphocytes},
author = {Romagnoli, P. and Dooley, J. and Enault, G. and Vicente, R. and Malissen, B. and Liston, A. and van Meerwijk, J. P.M.},
url = {http://www.ncbi.nlm.nih.gov/pubmed/22988035},
doi = {10.4049/jimmunol.1201564},
issn = {1550-6606 (Electronic)
0022-1767 (Linking)},
year = {2012},
date = {2012-01-01},
journal = {Journal of Immunology},
volume = {189},
number = {8},
pages = {3831-7},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
|
2011
|
Pomie, C.; Vicente, R.; Vuddamalay, Y.; Ardesjö Lundgren, B.; van der Hoek, M.; Enault, G.; Kagan, J.; Fazilleau, N; Scott, H. S.; Romagnoli, P.; van Meerwijk, J. P. M. AIRE-deficient CD8+CD28low regulatory T lymphocytes fail to control experimental colitis Article de journal Dans: Proceedings of the National Academy of Sciences of the U.S.A., vol. 108, no. 30, p. 12437-12442, 2011. @article{RN162,
title = {AIRE-deficient CD8+CD28low regulatory T lymphocytes fail to control experimental colitis},
author = {Pomie, C. and Vicente, R. and Vuddamalay, Y. and Ardesjö Lundgren, B. and van der Hoek, M. and Enault, G. and Kagan, J. and Fazilleau, N and Scott, H. S. and Romagnoli, P. and van Meerwijk, J.P.M.},
year = {2011},
date = {2011-01-01},
journal = {Proceedings of the National Academy of Sciences of the U.S.A.},
volume = {108},
number = {30},
pages = {12437-12442},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
|
Pasquet, Lise; Joffre, Olivier; Santolaria, Thibault; van Meerwijk, Joost Hematopoietic chimerism and transplantation tolerance: a role for regulatory T cells Article de journal Dans: Frontiers in Immunology, vol. 2, 2011, ISSN: 1664-3224. @article{RN163,
title = {Hematopoietic chimerism and transplantation tolerance: a role for regulatory T cells},
author = {Pasquet, Lise and Joffre, Olivier and Santolaria, Thibault and van Meerwijk, Joost},
url = {http://www.frontiersin.org/Journal/Abstract.aspx?s=1180&name=immunological_tolerance&ART_DOI=10.3389/fimmu.2011.00080},
doi = {10.3389/fimmu.2011.00080},
issn = {1664-3224},
year = {2011},
date = {2011-01-01},
journal = {Frontiers in Immunology},
volume = {2},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
|
Nouzé, C.; Pasquet, L.; van Meerwijk, J. P. M. In vitro expansion of alloantigen-specific regulatory T cells and their use in prevention of allograft-rejection Chapitre d'ouvrage Dans: Kassiotis, G.; Liston, A. (Ed.): Regulatory T-Cells: Methods and Protocols, vol. 707, p. 187-196, Springer, 2011. @inbook{RN157,
title = {In vitro expansion of alloantigen-specific regulatory T cells and their use in prevention of allograft-rejection},
author = {Nouzé, C. and Pasquet, L. and van Meerwijk, J. P. M.},
editor = {Kassiotis, G. and Liston, A.},
year = {2011},
date = {2011-01-01},
booktitle = {Regulatory T-Cells: Methods and Protocols},
volume = {707},
pages = {187-196},
publisher = {Springer},
series = {Methods in Molecular Biology},
keywords = {},
pubstate = {published},
tppubtype = {inbook}
}
|
2010
|
Romagnoli, P.; van Meerwijk, J. P. M. Thymic selection and lineage commitment of CD4+Foxp3+ regulatory T lymphocytes Article de journal Dans: Prog Mol Biol Transl Sci., vol. 92, p. 251-77, 2010. @article{RN156,
title = {Thymic selection and lineage commitment of CD4+Foxp3+ regulatory T lymphocytes},
author = {Romagnoli, P. and van Meerwijk, J. P. M.},
year = {2010},
date = {2010-01-01},
journal = {Prog Mol Biol Transl Sci.},
volume = {92},
pages = {251-77},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
|
2008
|
Joffre, O.; Santolaria, T.; Calise, D.; Al Saati, T.; Hudrisier, D.; Romagnoli, P.; van Meerwijk, J. P. M. Prevention of acute and chronic allograft rejection with CD4+CD25+Foxp3+ regulatory T lymphocytes Article de journal Dans: Nature Medicine, vol. 14, no. 1, p. 88-92, 2008. @article{RN137,
title = {Prevention of acute and chronic allograft rejection with CD4+CD25+Foxp3+ regulatory T lymphocytes},
author = {Joffre, O. and Santolaria, T. and Calise, D. and Al Saati, T. and Hudrisier, D. and Romagnoli, P. and van Meerwijk, J. P. M.},
year = {2008},
date = {2008-01-01},
journal = {Nature Medicine},
volume = {14},
number = {1},
pages = {88-92},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
|
Romagnoli, P.; Ribot, J.; Tellier, J.; van Meerwijk, J. P. M. Thymic and peripheral generation of CD4+Foxp3+ regulatory T cells Chapitre d'ouvrage Dans: Jiang, S. (Ed.): Regulatory T cells and clinical application, vol. in press, Springer, 2008, ISBN: 978-0-387-77908-9. @inbook{RN134,
title = {Thymic and peripheral generation of CD4+Foxp3+ regulatory T cells},
author = {Romagnoli, P. and Ribot, J. and Tellier, J. and van Meerwijk, J. P. M.},
editor = {Jiang, S.},
url = {http://www.springer.com/biomed/immunology/book/978-0-387-77908-9},
isbn = {978-0-387-77908-9},
year = {2008},
date = {2008-01-01},
booktitle = {Regulatory T cells and clinical application},
volume = {in press},
publisher = {Springer},
keywords = {},
pubstate = {published},
tppubtype = {inbook}
}
|
Joffre, O.; Santolaria, T.; van Meerwijk, J. P. M. [Tregs-based immunotherapy: an efficient way to fully inhibit acute and chronic rejection.] Article de journal Dans: Med Sci (Paris), vol. 24, no. 8-9, p. 689-691, 2008, ISSN: 0767-0974 (Print). @article{RN153,
title = {[Tregs-based immunotherapy: an efficient way to fully inhibit acute and chronic rejection.]},
author = {Joffre, O. and Santolaria, T. and van Meerwijk, J.P.M.},
url = {http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=18789210},
doi = {00/00/0C/B6/ [pii]},
issn = {0767-0974 (Print)},
year = {2008},
date = {2008-01-01},
journal = {Med Sci (Paris)},
volume = {24},
number = {8-9},
pages = {689-691},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
|
Pomie, C.; Menager-Marcq, I.; van Meerwijk, J. P. M. Murine CD8(+) regulatory T lymphocytes: The new era Article de journal Dans: Hum Immunol, vol. 69, p. 708-714, 2008, ISSN: 0198-8859 (Print). @article{RN155,
title = {Murine CD8(+) regulatory T lymphocytes: The new era},
author = {Pomie, C. and Menager-Marcq, I. and van Meerwijk, J.P.M.},
url = {http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=18817827},
doi = {S0198-8859(08)00450-3 [pii]
10.1016/j.humimm.2008.08.288},
issn = {0198-8859 (Print)},
year = {2008},
date = {2008-01-01},
journal = {Hum Immunol},
volume = {69},
pages = {708-714},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
|
2007
|
Ribot, J.; Enault, G.; Pilipenko, S.; Huchencq, A.; Calise, M.; Hudrisier, D.; Romagnoli, P.; van Meerwijk, J. P. M. Shaping of the autoreactive regulatory T cell repertoire by thymic cortical positive selection Article de journal Dans: J. Immunol., vol. 179, p. 6741-6748, 2007. @article{RN135,
title = {Shaping of the autoreactive regulatory T cell repertoire by thymic cortical positive selection},
author = {Ribot, J. and Enault, G. and Pilipenko, S. and Huchencq, A. and Calise, M. and Hudrisier, D. and Romagnoli, P. and van Meerwijk, J. P. M.},
year = {2007},
date = {2007-01-01},
journal = {J. Immunol.},
volume = {179},
pages = {6741-6748},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
|
2006
|
Menager-Marcq, I.; Pomie, C.; Romagnoli, P.; van Meerwijk, J. P. M. CD8+CD28- regulatory T lymphocytes prevent experimental inflammatory bowel disease in mice Article de journal Dans: Gastroenterology, vol. 131, no. 6, p. 1775-85, 2006. @article{RN132,
title = {CD8+CD28- regulatory T lymphocytes prevent experimental inflammatory bowel disease in mice},
author = {Menager-Marcq, I. and Pomie, C. and Romagnoli, P. and van Meerwijk, J. P. M.},
url = {http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=17087950},
year = {2006},
date = {2006-01-01},
journal = {Gastroenterology},
volume = {131},
number = {6},
pages = {1775-85},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
|
Tellier, J.; van Meerwijk, J. P. M.; Romagnoli, P. An MHC-linked locus modulates thymic differentiation of CD4+CD25+Foxp3+ regulatory T lymphocytes Article de journal Dans: Int Immunol, vol. 18, no. 11, p. 1509-1519, 2006. @article{RN133,
title = {An MHC-linked locus modulates thymic differentiation of CD4+CD25+Foxp3+ regulatory T lymphocytes},
author = {Tellier, J. and van Meerwijk, J. P. M. and Romagnoli, P.},
url = {http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=16943258},
year = {2006},
date = {2006-01-01},
journal = {Int Immunol},
volume = {18},
number = {11},
pages = {1509-1519},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
|
Ribot, J.; Romagnoli, P.; van Meerwijk, J. P. M. Agonist ligands expressed by thymic epithelium enhance positive selection of regulatory T lymphocytes from precursors with a normally diverse TCR-repertoire Article de journal Dans: J. Immunol., vol. 177, no. 2, p. 1101-1107, 2006. @article{RN128,
title = {Agonist ligands expressed by thymic epithelium enhance positive selection of regulatory T lymphocytes from precursors with a normally diverse TCR-repertoire},
author = {Ribot, J. and Romagnoli, P. and van Meerwijk, J. P. M.},
year = {2006},
date = {2006-01-01},
journal = {J. Immunol.},
volume = {177},
number = {2},
pages = {1101-1107},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
|
Joffre, O.; van Meerwijk, J. P. M. CD4+CD25+ regulatory T lymphocytes in bone marrow transplantation Article de journal Dans: Seminars in Immunology, vol. 18, no. 2, p. 128-135, 2006. @article{RN127,
title = {CD4+CD25+ regulatory T lymphocytes in bone marrow transplantation},
author = {Joffre, O. and van Meerwijk, J. P. M.},
year = {2006},
date = {2006-01-01},
journal = {Seminars in Immunology},
volume = {18},
number = {2},
pages = {128-135},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
|
2005
|
Romagnoli, P.; Tellier, J.; van Meerwijk, J. P. M. Genetic control of thymic development of CD4+CD25+FoxP3+ regulatory T lymphocytes Article de journal Dans: Eur J Immunol, vol. 35, p. 3525-3532, 2005. @article{RN123,
title = {Genetic control of thymic development of CD4+CD25+FoxP3+ regulatory T lymphocytes},
author = {Romagnoli, P. and Tellier, J. and van Meerwijk, J. P. M.},
year = {2005},
date = {2005-01-01},
journal = {Eur J Immunol},
volume = {35},
pages = {3525-3532},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
|
Romagnoli, P.; Hudrisier, D.; van Meerwijk, J. P. M. Molecular signature of recent thymic selection events on effector and regulatory CD4+ T lymphocytes Article de journal Dans: Journal of Immunology, vol. 175, p. 5751-5758, 2005. @article{RN121,
title = {Molecular signature of recent thymic selection events on effector and regulatory CD4+ T lymphocytes},
author = {Romagnoli, P. and Hudrisier, D. and van Meerwijk, J. P. M.},
year = {2005},
date = {2005-01-01},
journal = {Journal of Immunology},
volume = {175},
pages = {5751-5758},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
|
2004
|
Joffre, O.; Gorsse, N.; Romagnoli, P.; Hudrisier, D.; van Meerwijk, J. P. M. Induction of antigen-specific tolerance to bone-marrow allografts with CD4+CD25+ T lymphocytes Article de journal Dans: Blood, vol. 103, no. 11, p. 4216-4221, 2004. @article{RN118,
title = {Induction of antigen-specific tolerance to bone-marrow allografts with CD4+CD25+ T lymphocytes},
author = {Joffre, O. and Gorsse, N. and Romagnoli, P. and Hudrisier, D. and van Meerwijk, J. P.M.},
url = {file://D:%5CJoost's%20files%5Cmanuscripts%5CBlood%202004%20(Treg%20BM%20transplantation)%5CBlood%20submission%5Cresubmission%5CJoffre%20et%20al.pdf},
year = {2004},
date = {2004-01-01},
journal = {Blood},
volume = {103},
number = {11},
pages = {4216-4221},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
|
Cannarile, M. A.; Decanis, N.; van Meerwijk, J. P.; Brocker, T. The role of dendritic cells in selection of classical and nonclassical CD8+ T cells in vivo Article de journal Dans: J Immunol, vol. 173, no. 8, p. 4799-805, 2004. @article{RN120b,
title = {The role of dendritic cells in selection of classical and nonclassical CD8+ T cells in vivo},
author = {Cannarile, M. A. and Decanis, N. and van Meerwijk, J. P. and Brocker, T.},
url = {file://localhost/Users/Joost/Documents/Lab/articles/Cannarile%20JI%202004.pdf},
year = {2004},
date = {2004-01-01},
journal = {J Immunol},
volume = {173},
number = {8},
pages = {4799-805},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
|
2003
|
Hudrisier, D.; Feau, S.; Bonnet, V.; Romagnoli, P.; van Meerwijk, J. P. M. In vivo maintenance of T lymphocyte unresponsiveness induced by thymic medullary epithelium requires antigen presentation by radioresistant cells Article de journal Dans: Immunology, vol. 108, p. 24-31, 2003. @article{RN108b,
title = {In vivo maintenance of T lymphocyte unresponsiveness induced by thymic medullary epithelium requires antigen presentation by radioresistant cells},
author = {Hudrisier, D. and Feau, S. and Bonnet, V. and Romagnoli, P. and van Meerwijk, J.P.M.},
year = {2003},
date = {2003-01-01},
journal = {Immunology},
volume = {108},
pages = {24-31},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
|
Cemerski, S.; van Meerwijk, J. P. M.; Romagnoli, P. Oxidative stress-induced T lymphocyte hyporesponsiveness is caused by structural modification rather than proteasomal degradation of crucial signaling molecules Article de journal Dans: Eur J Immunol, vol. 33, no. 8, p. 2178-85, 2003. @article{RN115,
title = {Oxidative stress-induced T lymphocyte hyporesponsiveness is caused by structural modification rather than proteasomal degradation of crucial signaling molecules},
author = {Cemerski, S. and van Meerwijk, J.P.M. and Romagnoli, P.},
year = {2003},
date = {2003-01-01},
journal = {Eur J Immunol},
volume = {33},
number = {8},
pages = {2178-85},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
|
Capone, M.; Lees, R. L.; Finke, D.; Ernst, B.; van Meerwijk, J. P. M.; MacDonald, H. R. Selective absence of CD8+ TCRalpha beta+ intestinal epithelial cells in transgenic mice expressing beta2-microglobulin-associated ligands exclusively on thymic cortical epithelium. Article de journal Dans: Eur J Immunol, vol. 33, no. 6, p. 1471-7, 2003. @article{RN114,
title = {Selective absence of CD8+ TCRalpha beta+ intestinal epithelial cells in transgenic mice expressing beta2-microglobulin-associated ligands exclusively on thymic cortical epithelium.},
author = {Capone, M. and Lees, R.L. and Finke, D. and Ernst, B. and van Meerwijk, J. P. M. and MacDonald, H. R.},
url = {D:Joost's filesarticlesCapone EJI 2003.pdf},
year = {2003},
date = {2003-01-01},
journal = {Eur J Immunol},
volume = {33},
number = {6},
pages = {1471-7},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
|
2002
|
Romagnoli, P.; Hudrisier, D.; van Meerwijk, J. P. M. Preferential recognition of self-antigens despite normal thymic deletion of CD4+CD25+ regulatory T cells Article de journal Dans: Journal of Immunology, vol. 168, p. 1644-1648, 2002. @article{RN103,
title = {Preferential recognition of self-antigens despite normal thymic deletion of CD4+CD25+ regulatory T cells},
author = {Romagnoli, P. and Hudrisier, D. and van Meerwijk, J.P.M.},
url = {D:Joost's filesarticlesRomagnoli JI 2002.pdf},
year = {2002},
date = {2002-01-01},
journal = {Journal of Immunology},
volume = {168},
pages = {1644-1648},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
|
Constantin, A.; Lauwers-Cances, V.; Navaux, F.; Abbal, M.; van Meerwijk, J.; Mazieres, B.; Cambon-Thomsen, A.; Cantagrel, A. Stromelysin 1 (matrix metalloproteinase 3) and HLA-DRB1 gene polymorphisms: Association with severity and progression of rheumatoid arthritis in a prospective study Article de journal Dans: Arthritis Rheum, vol. 46, no. 7, p. 1754-62, 2002. @article{RN125,
title = {Stromelysin 1 (matrix metalloproteinase 3) and HLA-DRB1 gene polymorphisms: Association with severity and progression of rheumatoid arthritis in a prospective study},
author = {Constantin, A. and Lauwers-Cances, V. and Navaux, F. and Abbal, M. and van Meerwijk, J. and Mazieres, B. and Cambon-Thomsen, A. and Cantagrel, A.},
url = {http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=12124858},
year = {2002},
date = {2002-01-01},
journal = {Arthritis Rheum},
volume = {46},
number = {7},
pages = {1754-62},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
|
Constantin, A.; Lauwers-Cances, V.; Navaux, F.; Abbal, M.; van Meerwijk, J.; Mazieres, B.; Cambon-Thomsen, A.; Cantagrel, A. Collagenase-1 (MMP-1) and HLA-DRB1 gene polymorphisms in rheumatoid arthritis: a prospective longitudinal study Article de journal Dans: J Rheumatol, vol. 29, no. 1, p. 15-20, 2002. @article{RN126,
title = {Collagenase-1 (MMP-1) and HLA-DRB1 gene polymorphisms in rheumatoid arthritis: a prospective longitudinal study},
author = {Constantin, A. and Lauwers-Cances, V. and Navaux, F. and Abbal, M. and van Meerwijk, J. and Mazieres, B. and Cambon-Thomsen, A. and Cantagrel, A.},
url = {http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=11824952},
year = {2002},
date = {2002-01-01},
journal = {J Rheumatol},
volume = {29},
number = {1},
pages = {15-20},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
|
Cemerski, S.; Cantagrel, A.; van Meerwijk, J. P. M.; Romagnoli, P. Reactive oxygen species differentially affect T cell receptor signaling pathways Article de journal Dans: Journal of Biological Chemistry, vol. 277, p. 19585-19593, 2002. @article{RN106,
title = {Reactive oxygen species differentially affect T cell receptor signaling pathways},
author = {Cemerski, S. and Cantagrel, A. and van Meerwijk, J. P.M. and Romagnoli, P.},
url = {D:Joost's filesarticlesCemerski JBC 2002.pdf},
year = {2002},
date = {2002-01-01},
journal = {Journal of Biological Chemistry},
volume = {277},
pages = {19585-19593},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
|
2001
|
Romagnoli, P.; Strahan, D.; Pelosi, M.; Cantagrel, A.; van Meerwijk, J. P. M. A potential role for tyrosine kinase p56lck in rheumatoid arthritis synovial fluid T lymphocyte hyporesponsiveness Article de journal Dans: International Immunology, vol. 13, no. 3, p. 305-312, 2001. @article{RN91,
title = {A potential role for tyrosine kinase p56lck in rheumatoid arthritis synovial fluid T lymphocyte hyporesponsiveness},
author = {Romagnoli, P. and Strahan, D. and Pelosi, M. and Cantagrel, A. and van Meerwijk, J.P.M.},
year = {2001},
date = {2001-01-01},
journal = {International Immunology},
volume = {13},
number = {3},
pages = {305-312},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
|
 Le répertoire des lymphocytes T régulateurs est auto-spécifique
Le répertoire des lymphocytes T régulateurs est auto-spécifique